Abstract
Background:
Dermatophytes are a group of keratinophilic fungi worldwide, which can infect the skin, hair and nails of humans and animals. This genus includes several species that present different features of dermatophytosis. Although, laboratory diagnosis of dermatophytes is based on direct microscopy, biochemical tests and culture, these manners are expensive, time consuming and need skilled staff. Therefore, molecular methods like PCR-RFLP are the beneficial tools for identification, which are rapid and sensitive. Thus, dermatophyte species are able to generate characteristic band patterns on agarose gel electrophoresis using PCR-RFLP technique, which leads to successful identification at the species level within a 5-hour period.Objectives:
The purpose of this study was to study inter- and intraspecific genomic variations for identification of clinically important dermatophyte species obtained from clinical specimens in Isfahan, Iran using PCR-RFLP.Materials and Methods:
From March 2011 to August 2012, 135 clinical isolates were collected from infected patients at Isfahan, Iran. ITS1-5.8S-ITS2 region of rDNA was amplified using universal fungal primers. Subsequently, amplified products were digested by the MvaI restriction enzyme. Using discriminating band profiles on agarose gel, dermatophyte species were identified. However, DNA sequencing was used for unidentifiable strains.Results:
The specimens were obtained from skin scrapings (70.3%), nail (24.4%) and hair (5.1%) clippings. Most patients were between 21 - 30 years and the ratio of male to female was 93/42. Trichophyton interdigitale was the commonest isolate (52.5%) in our findings, followed by Epidermophyton floccosum (24.4%), T. rubrum (16.2%), Microsporumcanis (2.2%), T. erinacei (1.4%), T. violaceum (1.4%), T. tonsurans (0.7%) and M. gypseum (0.7%) based on PCR-RFLP.Conclusions:
Combination of traditional methods and molecular techniques considerably improves identification of dermatophytes in the species level in clinical laboratories, which can lead to properly antifungal therapy and successful management of infections. However, restriction and specificity and sensitivity should be lowered and increased, respectively, to be useful for a wide variety of clinical applications.Keywords
1. Background
Dermatophytosis caused by the genus of dermatophytes taxonomically classified into three genera (Epidermophyton, Microsporum and Trichophyton) is thought to be one of the most significant public health problems yet not solved. Dermatophytes are highly specialized pathogenic fungi and the most common cause of superficial mycoses in humans and animals, affecting millions of individuals annually (1). The estimated lifetime risk of acquiring a dermatophyte infection is 10 - 20%. They have the capacity to invade keratinized tissue to produce infections that are generally restricted to the corneocytes of the skin, hair and nails due to host defense reactions in immunocompetent individuals; however, dermatophytosis in immunocompromised hosts, i.e. severe burns, diabetes and malignancies may progress to subcutaneous and deep cutaneous disorder (2, 3). Poor hygienic conditions, over-population and a highly humid weather are causative factors of dermatophytosis.
Trichophyton rubrum, T. mentagrophytes, Microsporum canis and Epidermophyton floccosum are distributed worldwide, but some species such as M. audouinii (Africa), T. violaceum (Africa, Asia and Europe), T. soudanense (Africa) and T. tonsurans (Americas and Europe) have geographical restrictions (4, 5). Laboratory diagnosis of dermatomycosis is based on demonstration of hyphae and both macro- and micro-conidia, by direct microscopic examination of clinical samples, followed by biochemical characteristics and culture. Important characteristics are the rate of growth, shape and texture of the culture on solid media, color, diffusion of pigments into the agar and sporulation (6, 7). However, this system of identification is time-consuming and may pose difficulties for non-experts in differentiation of the morphology of cultured colonies. Furthermore, even the same strains may show morphologically diverse colonies, making the identification of the causative organism more difficult. Moreover, the phenotypic features can be easily influenced by outside factors such as temperature variation, medium and chemotherapy (8).
Due to the high degree of phenotypic similarity between dermatophyte species, identification problems are imminent. Conventional approaches for identification down to the species level in the diagnostic laboratory are based on morphological and physiological criteria, need several days or weeks to be concluded and are frequently unspecific. However, today molecular identification methods are well established. Sequencing of the partial ribosomal operon is relatively expensive. Therefore, alternative molecular tools with sufficient specificity, reproducibility and sensitivity are necessary. Recently, molecular approach like restriction fragment length polymorphism (RFLP) is a technique based on detection of genomic restriction fragments by PCR amplification, which can be used with DNA of any organism and has been proven to be useful for rapid and correct identification of dermatophyte species and able to generate species-specific DNA polymorphisms with many dermatophyte species on the basis of characteristic band patterns detected by agarose gel electrophoresis (9). In particular, since the relatively invariant marker gene internal transcribed spacer (ITS rDNA) regions are used as standard in dermatophytes have shown promise as targets for identification at the species level and can be used in successful identification (10, 11).
2. Objectives
The purpose of the present paper was to study inter- and intraspecific genomic variations for identification of clinically important dermatophyte species obtained from clinical specimens in Isfahan, Iran using PCR-RFLP.
3. Materials and Methods
The isolates used in the current study were obtained from patients suspected to dermatophytosis and handled under biosafety level II conditions. A total of 167 clinical specimens recovered from different origins were initially examined by direct examination using 20% potassium hydroxide (KOH) to examine macro/micro conidia and the shape of hyphae, subsequently inoculated on Sabouraud’s dextrose agar supplemented with chloramphenicol (SC) (Difco, USA) and cycloheximide (SCC) (Difco, USA) at 30°C for four weeks. Then, strain identities were verified to the species level using PCR- RFLP based on internal transcribed spacer regions of rDNA (ITS rDNA).
DNA Extraction: DNA extraction was previously described (12, 13); briefly, mycelia were grown on 2% Malt extract agar (Difco, USA) for 2 weeks at 27°C. A sterile blade was used to scrape off the biomass from the surface of plate and transferred to a 2 mL Eppendorf tube containing 300 μL lysis buffer (200 mM Tris/HCl with pH = 7.5, 25 mM EDTA, 0.5% SDS, 250 mM NaCl). Then cells were mechanically disrupted for approximately one minute and incubated at 100°C for 15 minutes, subsequently 150 mL of 3.0 M sodium acetate buffer was added, the mixture was vortexed and incubated for 10 minutes at -20°C, the solution was mixed and centrifuged for 5 minutes at 10000 rpm. The supernatant transferred to a new tube and extracted with phenol/chloroform (1:1, v/v). DNA was allowed to precipitate with an equal volume of isopropanol for 10 minutes at -20°C and then centrifuged for 10 minutes at 12000 rpm. The pellets were washed with cold 70% ethanol, dried at room temperature, resuspended in 97.5 mL of TE-buffer with 2.5 mL of RNase 20 U/mL and incubated for 5 minutes at 37°C. DNA extracts were stored at -20°C prior to use.
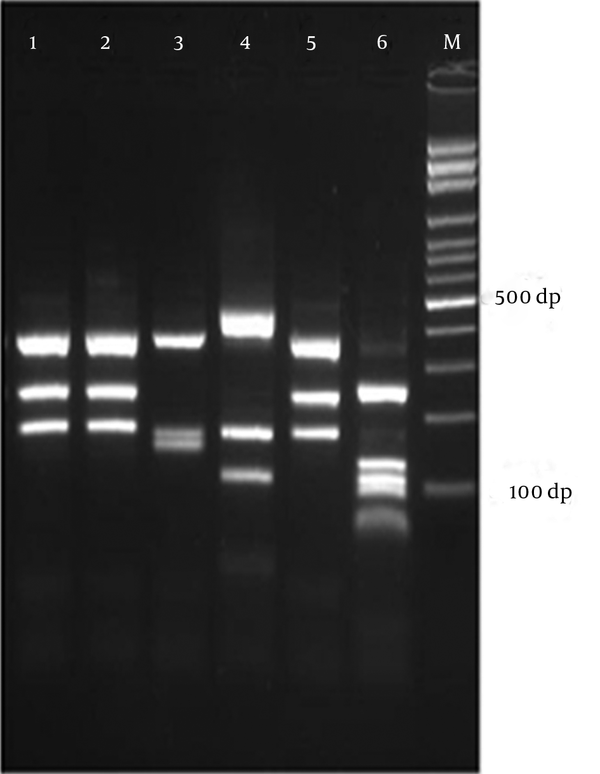
Molecular identification: ITS rDNA region was amplified using universal fungal primers ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) (Sina Gene, Iran) (14). Briefly, PCR reactions were performed on a Gene Amp PCR System 9700 (Applied Biosystems, Foster City, CA) in 25 μL volumes containing 2 μL of template DNA, 2.5 μL of 10 × reaction buffer (0.1 M Tris/HCl, pH 8.0, 0.5 M KCl, 1.5 mM MgCl2, 0.1% gelatine, 1% Triton X-100), 0.4 mM of each dNTP, 30 pmol of each forward and reverse primers and 1.25 U Taq DNA polymerase (Amplicon, Denmark). Amplification of ITS rDNA was performed with cycles of 5 minutes at 94°C for primary denaturation, followed by 40 cycles at 94°C (30 seconds), 58°C (30 seconds) and 72°C (60 seconds), with a final 7-minute extension step at 72°C. RFLP procedures were as follow; 10 μL of amplified products (ITS rDNA) added to 1.5 μL of 10× buffer, 0.5 μL (5U) of the MvaI restriction enzyme (Fast digest; Fermentas, Vilnius, Lithuania) which recognizes the sequence 5’ CC (T/A) GG 3’ (14), and 3 μL of double distilled water. Subsequently reactions were incubated at 37°C for 15 minutes and then the electrophoresis was performed using TBE buffer (Tris 0.09 M, Boric acid 0.09 M, EDTA 2mM), at 80V for 90 minutes. Five microliter of PCR amplicons and 12 μL of each RFLP products were loaded on 1.5 and 2% agarose gel, respectively, and stained with 0.5 μg/mL ethidium bromide. The bands on agarose gel were visualized using gel documentation system and photographed.
Sequence analysis of ITS rDNA: To confirm the reliability of molecular methods used for identification of dermatophyte species, we performed DNA sequencing of ITS rDNA region randomly for five unknown species. PCR products were purified using GFX PCR DNA (GE Healthcare, Buckinghamshire, UK). Sequencing was performed using big dye terminator chemistry as described in the manufacture’s manual. Sequence data obtained was adjusted using Lasergene SeqMan software version 9.0.4 (DNAStar, Madison, Wisconsin, USA) and compared with GenBank. The sequence of the ITS rDNA region for dermatophyte species determined in the present study deposited in GenBank with the accession numbers from KF437398 to KF437402.
4. Results
The study group comprised 167 patients attending to the mycological laboratory of Isfahan University of Medical Sciences, Isfahan, Iran, which clinically suspected to dermatophytosis collected over the one year study period, from March 2011 until August 2012. Due to mycological and molecular criteria, 135 of 167 patients were identified as dermatophytosis due to different species. Table 1 summarizes data of all infected patients listed by percentage of different isolated species. In contrast, the rest of patients had positive results for dermatomycosis due to different genera like Aspergillus spp., Fusaraium spp., Candida spp. and Chrysosporium spp. The specimens were obtained from skin scrapings (n = 95; 70.3%), nail (n = 34; 25.1%) and hair (n = 7; 5.1%) clippings. The most common clinical presentation among skin dermatophytosis was tinea pedis (n = 42; 44.2%), followed by tinea corporis (n = 20; 21%), tinea cruris (n = 19; 20%), tinea manuum (n = 11; 11.5%) and tinea faciei (n = 2; 2.1%).
Dermatophyte Species Isolated From Clinical Specimens in Isfahan, Iran, From March 2011 Until August 2012 a
| Species | Tinea pedis | Tinea Unguium | Tinea Corporis | Tinea Cruris | Tinea Manuum | Tinea Capitis | Tinea Faciei | Total Number | Ratio of Male to Female (M/F) |
|---|---|---|---|---|---|---|---|---|---|
| T. interdigitale | 31 (72) | 12 (36.3) | 13 (65) | 5 (26.3) | 8 (72.7) | 2 (28.5) | 0 (0) | 71 (52.5) | 53/18 |
| E. floccosum | 3 (6.9) | 10 (30.3) | 4 (20) | 14 (73.6) | 2 (18.1) | 0 (0) | 0 (0) | 33 (24.4) | 20/13 |
| T. rubrum | 7 (16.2) | 11 (33.3) | 2 (10) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 22 (16.2) | 15/7 |
| M. canis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (42.8) | 0 (0) | 3 (2.2) | 2/1 |
| T. erinacei | 1 (2.3) | 0 (0) | 0 (0) | 0 (0) | 1 (9) | 0 (0) | 0 (0) | 2 (1.4) | 0/2 |
| T. violaceum | 0 (0) | 1 (2.3) | 0 (0) | 0 (0) | 0 (0) | 1 (14.2) | 0 (0) | 2 (1.4) | 1/1 |
| T. tonsurans | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (14.2) | 0 (0) | 1 (0.7) | 1/0 |
| M. gypseum | 0 (0) | 0 (0) | 1 (5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0.7) | 1/0 |
| Total number | 42 (31.1) | 34 (25.1) | 20 (14.8) | 19 (14) | 11 (8.1) | 7 (5.1) | 2 (1.4) | 135 (100) | 135 |
Molecular analysis (RFLP assay; Figure 1) confirmed the results and showed that T.interdigitale was the commonest isolate (52.5%), followed by E. floccosum (24.4%), T. rubrum (16.3%), M. canis (2.2%), T. erinacei (1.4%), T. violaceum (1.4%), T. tonsurans (0.7%) and M.gypseum (0.7%) (Table 1). Most patients aged 21 - 30 years (Table 2) and the ratio of male to female in different groups were: T. interdigitale: 53/18, E. floccosum: 20/13, T. rubrum: 15/7, M. canis: 2/1, T. erinacei: 0/2, T. violaceum: 1/1, T. tonsurans: 1/0, M. gypseum: 1/0. T. violaceum isolates were obtained from nail (Tinea unguium) and hair (Tinea capitis = endothrix). All isolated strains of M. canis and T. tonsurans were originated from hair (ectothrix/endothrix and invasion, respectively); however, M. gypseum was isolated from skin scarping.
Electrophoretic Profile of ITS-RFLP with MvaI for Some Dermatophyte Species, Obtained From Clinical Specimens in Isfahan, Iran
Age Range of Patients With Dermatophytosis Isolated From Isfahan, Iran
| Bracket age | Dermatophyte Species | Total No. (%) |
|---|---|---|
| 0 - 10 | T. interdigitale (n = 4), T. rubrum (n = 4), M. canis (n = 2) | 10 (7.4) |
| 11 - 20 | T. interdigitale (n = 7), T. rubrum (n = 4), E. floccosum (n = 2), M. canis (n = 1) | 14 (10.3) |
| 21 - 30 | T. interdigitale (n = 12), E. floccosum (n = 10), T. rubrum (n = 6), T. tonsurans (n = 1), T. violaceum (n = 1) | 30 (22.2) |
| 31 - 40 | T. interdigitale (n = 11), T. rubrum (n = 2), E. floccosum (n = 2), M. gypseum (n = 1) | 16 (11.8) |
| 41 - 50 | T. interdigitale (n = 12), E. floccosum (n = 8), T. rubrum (n = 3), T. violaceum (n = 1), T. erinacei (n = 1) | 25 (18.5) |
| 51 - 60 | T. interdigitale (n = 13), E. floccosum (n = 3), T. erinacei (n = 1) | 17 (12.5) |
| 61 - 70 | T. interdigitale (n = 9), E. floccosum (n = 4), T. rubrum (n = 3) | 16 (11.8) |
| 71 - 80 | E. floccosum (n = 4), T. interdigitale (n = 3) | 7 (5.1) |
Surprisingly, in the current study we described the first case of tinea pedis in Iran and tinea manuum due to T. erinacei in a 45-year-old female and 60-year-old female, respectively. These isolates produced yellowish, flat, powdery and granular colonies on SCC. In direct microscopic examination of these strains, a great number of large clavate microconidia were born on the sides of hyphae and rarely, 2 - 3 celled macro-conidia (shorter than those seen in T. interdigitale), were observed. PCR-RFLP analysis using a single restriction enzyme like MvaI differentiated all dermatophyte species with different band patterns; although, some species like T. interdigitale, T. simii and M. gypseum have two band patterns and some of them such as M. canis/M. ferrugineum and T. equinum/T. tonsurans have the same profiles; therefore, powerful discriminative molecular tools such as DNA sequencing were used for unidentifiable strains. Therefore, ITS rDNA (ITS1-5.8S-ITS2) region sequencing identified T. rubrum (n = 2), T. erinacei (n = 2) and T. tonsurans (n = 1) (Table 3).
Five Suspicious Isolates Were Identified by Sequence Analysis in the Present Study
| Dermatophyte Species | Location Body | Gender | Age | Accession Number |
|---|---|---|---|---|
| T. rubrum | Skin | Male | 44 | KF437398 |
| T. rubrum | Nail | Male | 39 | KF437402 |
| T. erinacei | Hand | Female | 60 | KF437400 |
| T. erinacei | Foot | Female | 45 | KF437401 |
| T. tonsurans | Hair | Male | 27 | KF437399 |
5. Discussion
Although, routine identification of dermatophytes was accomplished by mycological examination of the clinical specimen (nail, skin, and hair) with potassium hydroxide followed by culturing, it is time-consuming and requires up to four weeks for the growth of organism and to observe typical features of the dermatophyte species directly from the clinical specimens. Therefore, correct identification in the species level, which can be important for prognosis and treatment, is crucially recommended. At this moment, ITS rDNA sequencing is the gold standard for identification of dermatophytes and relatives; the technique is relatively expensive, time-consuming and impractical for analysis of large numbers of isolates for epidemiological studies. In addition, the GenBank database is filled with incorrect sequences hampering identification by Blast; therefore, a rapid and validated method is urgently needed.
Recently, several PCR based techniques like arbitrarily primed PCR (AP-PCR) (15), random amplified polymorphic DNA (RAPD) (16, 17), repetitive sequence PCR (rep-PCR) (18), restriction analysis of the mitochondrial DNA (19, 20), semi-nested PCR (21), nested PCR (22), multiplex PCR (23) and single-strand conformation polymorphism (SSCP) analysis (24) are the available techniques for identification of dermatophytes. However, few methods reported a low sensitivity and specificity in identification of dermatophyte species. Thus, the current study was adjusted to use the PCR-RFLP based on internal transcribed spacer (ITS) region to rapidly identify isolated dermatophytes in the species level using restriction enzyme MvaI. Interestingly, Rezaei-Matehkolahei et al. studied molecular epidemiology of dermatophytosis in Tehran, Iran using RFLP analysis based on the ITS rDNA regions. They announced that tinea pedis was the most prevalent type of infection (43.4%) and T. interdigitale was the most common isolate, which are in accordance with the present study (31.8%) (25).
RFLP analysis with MvaI, shows two distinctive patterns for each T. interdigitale and M. gypseum (26). In our study, we found 50 (70%) T. interdigitale isolates with pattern I and 21(30%) isolates with pattern II and M. gypseum created band profile in accordance with pattern I. Kamiya et al. used PCR-RFLP technique targeting the DNA topoisomerase II gene for identification of 352 clinical isolates collected from patients with dermatophytosis and T. rubrum was the most prevalent isolated species; whereas, T. rubrum had the third place in our ranking (27). Similar to our study, they identified only one M. gypseum among isolates. There is a disagreement between our results and Falahati et al. outcomes (28). They reported E. floccosum as the most frequent dermatophyte species isolated in Tehran. They also reported Tinea corporis as the most common dermatophytosis; whereas, Tinea pedis was the most frequent infection in our findings.
Chadeganipour et al. showed that tinea capitis was the most prevalent clinical form (54.1%), followed by tinea corporis, tinea pedis (8.9%), tinea cruris (6.8%), tinea unguium (3.5%), tinea manuum (2.6%) and tinea barbae (0.3%). Predominant isolate in their study was T. verrucosum (32.8%) (29). From 1994 to 2001 Bassiri Jahromi and Khaksar studied 209 patients with tinea capitis and showed that T. violaceum was the most common causative agent; whereas, M. canis was the most widespread agent in patients with hair invasion in our experimentation (30). In investigations performed in Libya and Yemen, tinea corporis was the most frequent clinical form of dermatophytosis, but tinea pedis was recorded in only 10%, contradictory with our findings (31, 32).
Foster et al. showed that T. rubrum remained the most prevalent fungal pathogen in the United States during 1999-2002 (33). T. rubrum also ranked the first in Puerto Rico (85.7%) in a study performed by Vazquez and Sanchez on tinea corporis and tinea pedis (34). In their study, T. mentagrophytes accounted for 4% of total infections. Various publications indicate the high frequency of tinea pedis in different groups such as miners, butchers, sportsman, and soldiers suggesting the role of lifestyle (35-38). Our RFLP results are in line with previous sequencing data and show obvious differences among clinically important dermatophyte species as agents of dermatophytosis. On the basis of RFLP patterns, we did not observe any misidentification for those taxa. In spite of the fact that the incidence of tinea capitis is decreasing in developed countries, tinea pedis and onychomycosis are becoming an epidemiologic and economic complication. Therefore, distribution of dermatophyte species is considerably miscellaneous all around the world. Meticulous and rapid diagnosis of the genus and species of the dermatophyte using molecular techniques such as PCR-RFLP can lead to an appropriate antifungal therapy and more effective control of infections.
Acknowledgements
References
-
1.
Liu T, Zhang Q, Wang L, Yu L, Leng W, Yang J, et al. The use of global transcriptional analysis to reveal the biological and cellular events involved in distinct development phases of Trichophyton rubrum conidial germination. BMC Genomics. 2007;8:100. [PubMed ID: 17428342]. https://doi.org/10.1186/1471-2164-8-100.
-
2.
Garg J, Tilak R, Garg A, Prakash P, Gulati AK, Nath G. Rapid detection of dermatophytes from skin and hair. BMC Res Notes. 2009;2:60-65. [PubMed ID: 19374765]. https://doi.org/10.1186/1756-0500-2-60.
-
3.
Hryncewicz-Gwozdz A, Jagielski T, Dobrowolska A, Szepietowski JC, Baran E. Identification and differentiation of Trichophyton rubrum clinical isolates using PCR-RFLP and RAPD methods. Eur J Clin Microbiol Infect Dis. 2011;30(6):727-31. [PubMed ID: 21416216]. https://doi.org/10.1007/s10096-010-1144-3.
-
4.
Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008;51 Suppl 4:2-15. [PubMed ID: 18783559].
-
5.
Sahai S, Mishra D. Change in spectrum of dermatophytes isolated from superficial mycoses cases: first report from Central India. Indian J Dermatol Venereol Leprol. 2011;77(3):335-6. [PubMed ID: 21508578]. https://doi.org/10.4103/0378-6323.79718.
-
6.
Kim KH. Identification of dermatophytes. Korean J Med Mycol. 1997;2(1):1-8.
-
7.
Ninet B, Jan I, Bontems O, Lechenne B, Jousson O, Panizzon R, et al. Identification of dermatophyte species by 28S ribosomal DNA sequencing with a commercial kit. J Clin Microbiol. 2003;41(2):826-30. [PubMed ID: 12574293].
-
8.
Liu D, Coloe S, Baird R, Pedersen J. Application of PCR to the identification of dermatophyte fungi. J Med Microbiol. 2000;49(6):493-7. [PubMed ID: 10847201].
-
9.
Faggi E, Pini G, Campisi E, Bertellini C, Difonzo E, Mancianti F. Application of PCR to distinguish common species of dermatophytes. J Clin Microbiol. 2001;39(9):3382-5. [PubMed ID: 11526185].
-
10.
Graser Y, De Hoog S, Summerbell RC. Dermatophytes: recognizing species of clonal fungi. Med Mycol. 2006;44(3):199-209. [PubMed ID: 16702098]. https://doi.org/10.1080/13693780600606810.
-
11.
Summerbell RC, Moore MK, Starink-Willemse M, Van Iperen A. ITS barcodes for Trichophyton tonsurans and T. equinum. Med Mycol. 2007;45(3):193-200. [PubMed ID: 17464840]. https://doi.org/10.1080/13693780601087614.
-
12.
Makimura K, Tamura Y, Mochizuki T, Hasegawa A, Tajiri Y, Hanazawa R, et al. Phylogenetic classification and species identification of dermatophyte strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Clin Microbiol. 1999;37(4):920-4. [PubMed ID: 10074502].
-
13.
Liu D, Coloe S, Baird R, Pederson J. Rapid mini-preparation of fungal DNA for PCR. J Clin Microbiol. 2000;38(1):471. [PubMed ID: 10681211].
-
14.
Jackson CJ, Barton RC, Evans EG. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J Clin Microbiol. 1999;37(4):931-6. [PubMed ID: 10074504].
-
15.
Liu D, Coloe S, Pedersen J, Baird R. Use of arbitrarily primed polymerase chain reaction to differentiate Trichophyton dermatophytes. FEMS Microbiol Lett. 1996;136(2):147-50. [PubMed ID: 8869498].
-
16.
Howell SA, Barnard RJ, Humphreys F. Application of molecular typing methods to dermatophyte species that cause skin and nail infections. J Med Microbiol. 1999;48(1):33-40. [PubMed ID: 9920123].
-
17.
Baeza LC, Giannini MJ. Strain differentiation of Trichophyton rubrum by random amplification of polymorphic DNA (RAPD). Rev Inst Med Trop Sao Paulo. 2004;46(6):339-41. [PubMed ID: 15654481].
-
18.
Pounder JI, Williams S, Hansen D, Healy M, Reece K, Woods GL. Repetitive-sequence-PCR-based DNA fingerprinting using the Diversilab system for identification of commonly encountered dermatophytes. J Clin Microbiol. 2005;43(5):2141-7. [PubMed ID: 15872233]. https://doi.org/10.1128/JCM.43.5.2141-2147.2005.
-
19.
Mochizuki T, Takada K, Watanabe S, Kawasaki M, Ishizaki H. Taxonomy of Trichophyton interdigitale (Trichophyton mentagrophytes var. interdigitale) by restriction enzyme analysis of mitochondrial DNA. J Med Vet Mycol. 1990;28(3):191-6. [PubMed ID: 1698960].
-
20.
Kawasaki M, Aoki M, Ishizaki H, Nishio K, Mochizuki T, Watanabe S. Phylogenetic relationships of the genera Arthroderma and Nannizzia inferred from mitochondrial DNA analysis. Mycopathologia. 1992;118(2):95-102. [PubMed ID: 1359414].
-
21.
Yang G, Zhang M, Li W, An L. Direct species identification of common pathogenic dermatophyte fungi in clinical specimens by semi-nested PCR and restriction fragment length polymorphism. Mycopathologia. 2008;166(4):203-8. [PubMed ID: 18523862]. https://doi.org/10.1007/s11046-008-9130-3.
-
22.
Garg J, Tilak R, Singh S, Gulati AK, Garg A, Prakash P, et al. Evaluation of pan-dermatophyte nested PCR in diagnosis of onychomycosis. J Clin Microbiol. 2007;45(10):3443-5. [PubMed ID: 17699656]. https://doi.org/10.1128/JCM.02367-06.
-
23.
Kim JY, Choe YB, Ahn KJ, Lee YW. Identification of dermatophytes using multiplex polymerase chain reaction. Ann Dermatol. 2011;23(3):304-12. [PubMed ID: 21909200]. https://doi.org/10.5021/ad.2011.23.3.304.
-
24.
Cafarchia C, Otranto D, Weigl S, Campbell BE, Parisi A, Cantacessi C, et al. Molecular characterization of selected dermatophytes and their identification by electrophoretic mutation scanning. Electrophoresis. 2009;30(20):3555-64. [PubMed ID: 19862737]. https://doi.org/10.1002/elps.200900313.
-
25.
Rezaei-Matehkolaei A, Makimura K, de Hoog S, Shidfar MR, Zaini F, Eshraghian M, et al. Molecular epidemiology of dermatophytosis in Tehran, Iran, a clinical and microbial survey. Med Mycol. 2013;51(2):203-7. [PubMed ID: 22587730]. https://doi.org/10.3109/13693786.2012.686124.
-
26.
Rezaei-Matehkolaei A, Makimura K, Shidfar M, Zaini F, Eshraghian M, Jalalizand N, et al. Use of Single-enzyme PCR-restriction Digestion Barcode Targeting the Internal Transcribed Spacers (ITS rDNA) to Identify Dermatophyte Species. Iran J Public Health. 2012;41(3):82-94. [PubMed ID: 23113152].
-
27.
Kamiya A, Kikuchi A, Tomita Y, Kanbe T. PCR and PCR-RFLP techniques targeting the DNA topoisomerase II gene for rapid clinical diagnosis of the etiologic agent of dermatophytosis. J Dermatol Sci. 2004;34(1):35-48. [PubMed ID: 14757280].
-
28.
Falahati M, Akhlaghi L, Lari AR, Alaghehbandan R. Epidemiology of dermatophytoses in an area south of Tehran, Iran. Mycopathologia. 2003;156(4):279-87. [PubMed ID: 14682452].
-
29.
Chadeganipour M, Shadzi S, Dehghan P, Movahed M. Prevalence and aetiology of dermatophytoses in Isfahan, Iran. Mycoses. 1997;40(7-8):321-4. [PubMed ID: 9476518].
-
30.
Bassiri Jahromi S, Khaksar AA. Aetiological agents of tinea capitis in Tehran (Iran). Mycoses. 2006;49(1):65-7. [PubMed ID: 16367822].
-
31.
Ellabib MS, Khalifa Z, Kavanagh K. Dermatophytes and other fungi associated with skin mycoses in Tripoli, Libya. Mycoses. 2002;45(3-4):101-4. [PubMed ID: 12000510].
-
32.
Mahmoud AL. A study of dermatophytoses in Sana'a, Yemen Republic. Mycoses. 2002;45(3-4):105-8. [PubMed ID: 12000511].
-
33.
Foster KW, Ghannoum MA, Elewski BE. Epidemiologic surveillance of cutaneous fungal infection in the United States from 1999 to 2002. J Am Acad Dermatol. 2004;50(5):748-52. [PubMed ID: 15097959]. https://doi.org/10.1016/S0190.
-
34.
Vazquez M, Sanchez JL. A clinical and mycologic study of tinea corporis and pedis in Puerto Rico. Int J Dermatol. 1984;23(8):550-1. [PubMed ID: 6500796].
-
35.
Gotz H, Hantschke D. [A glance at the epidemiology of dermatomycoses in the coal mining industry]. Hautarzt. 1965;16(12):543-8. [PubMed ID: 5871099].
-
36.
Seebacher C, Bouchara JP, Mignon B. Updates on the epidemiology of dermatophyte infections. Mycopathologia. 2008;166(5-6):335-52. [PubMed ID: 18478365]. https://doi.org/10.1007/s11046-008-9100-9.
-
37.
Brocks KM, Johansen UB, Jorgensen HO, Ravnborg LR, Svejgaard EL. Tinea pedis and onychomycosis in Danish soldiers before and after service in ex-Yugoslavia. Mycoses. 1999;42(7-8):475-8. [PubMed ID: 10546489].
-
38.
Gentles JC, Evans EGV. Foot Infections in Swimming Baths. Br Med J. 1973;3(5874):260-2.