Abstract
Background:
Radio therapy plays an important role in controlling tumor growth in esophageal cancer patients.Objectives:
Our study provided comprehensive information about radio-sensitivity of Hdm2 gene in esophageal cancer cells in cancer cells.Methods:
The expression of Hdm2 gene in radiosensitive and radio-resistant esophageal squamous cell carcinoma TE1, TE8 and TE11 cell lines is studied. RNA extracted from each esophageal cancer cell, then Quantitative reverse transcription PCR (RT-qPCR) assay were used for measuring HDM2 gene expression. Finally, radiosensitivity was determined for every three esophageal cancer cell lines.Results:
It was found that Hdm2 gene significantly expressed in TE1, TE8 and TE11 cell lines by qRT-PCR analysis. Clonogenic assay showed that TE-11 was very sensitive to radiation relative to other cell lines.Conclusions:
Hdm2 protein binding to the P53 protein is a key regulator of cell cycle control and genome protection Cell cycle control, programmed cell death and response cells to radition play important roles.Keywords
Esophagus Cancer Hdm2 P53 Radiosensitivity Squamous Cell Carcinoma
1. Background
Esophageal cancer is the sixth deadly cancer with poor prognosis. The incidence and the prevalence rates are almost equal, and survival time is short (1). Squamous cell carcinoma (SCC) and adenocarcinoma could be assumed as predominant histology for patients who are suffering from esophageal carcinoma (2). More than 90% of esophageal cancers diagnosed in the worldwide have SCC origin. Small cell carcinoma has been seen in almost 1% of patients (3) and mukoepidermoid carcinoma in less than 1% (4). At the moment, despite the use of multimodality therapy, the 5-year survival rate is not even 50%. The utilization of auxiliary chemo-radiotherapy earlier revealed cancers with ESCCs, the most favorable result obtained for patients with high potential for lymph node metastasis (5-7). Radiotherapy plays a special role in tumor growth control in patients that are suffering from esophageal cancer. The new treatment method is assumed to ameliorate resection rates, decrease lymphatic metastases and increase survival time (6). Today chemo-radiotherapy is accepted as a standard method in management of locally advanced ESCCs without surgery. After the use of combination therapy survival rate improved significantly and recurrence decreased in patients with ESCCs (8). Esophageal cancer shows high radiosensitivity and it is assumed that the diversity in the pattern of gene expression is the main reason of diversities in the response of tumor cells to radiation (9).
Molecular analysis of esophageal cancer cells have revealed overexpression of some genes. Irrespective of the doubtful etiological factors and patient origin, genetic irregularity that frequently occur in SCC are: a) tumor suppressor genes Changes, distinctly p53, conducive to changed DNA repetition and correction, apoptosis and cell proliferation; b) the cell cycle’s control detriment and cut the G1/S cell cycle checkpoint; and c) changes in oncogene function conducting to deregulation of cell signaling cascades (10, 11). The p53 gene as a tumor suppressor gene induces apoptosis by up-regulating bax gene and also results in cell cycle arrest by down-regulating bcl-2 (12-14). Mdm2 in humans is called Hdm2 and it is a negative regulator for tumor suppressor p53 (2, 15). Hdm2 is overexpressed in various types of cancer, for example non-small cell lung cancer, melanoma esophageal cancer, breast cancer, leukemia, sarcoma and non-Hodgkin’s lymphoma (15). It is shown that a significant correlation between Mdm2 gene expression and p53 gene mutations. P53 missense mutations result in decreasing Mdm2 protein expression. In cancers with wild type (wt), p53 activity of p53 can be restored by inhibition of Hdm2; furthermore, in association with anti-tumor agents with apoptosis and inhibitory growth (1, 15). Extinguishing of Hdm2 mRNA have immediately elevated MCF-7 cell apoptosis and reduced the cell reproduction (2).
2. Objectives
In the present study, we reported the Hdm2 and P53 gene expression profile in theTE1, TE8 and TE11 ESCC cell lines that can illuminate the amount of radiosensitivity. The purpose of this study is investigation of the sensitivity of ESCC tumor cells as a function of hdm2 and P53 expression to irradiation.
3. Methods
3.1. Cell Lines
TE1 (RBRC- RCB1894), TE8 (RBRCRCB2098) and we prepared TE11 (RBRC-RCB2100) cell lines from RIKEN Bio-resource center (RIKEN; Tsukuba, Ibaraki, Japan). Then we cultured cells in RPMI1640 culture medium (Sigma-Aldrich, St. Louis, MO, US), and supplemented it at 37°C using 10% fetal bovine serum (FBS; Sigma, St. Louis, MO), 100 U/mL penicillin and 100 µg/ml streptomycin (Gibco, Inc., UK.) in a humidified incubator with 5% CO2.
3.2. RNA Extraction
RNX-Plus reagent was used for extracting the total RNA (Cinagen Co., Tehran, Iran). We treated the prepared cells (1 × 106 cells) using 1mL of RNX solution, at normal temperature we incubated it for 5 minutes, added a 200 µL chloroform, and centrifuged cell suspensions at 12000 RPM during 15 minutes at 4°C. We transferred the upper tack to another tube that was filled with equal volume of isopropanol and next we centrifuged it for 15 minutes at 4°C with high speed, supernatant discarded. Following this, RNA-pellet was washed by 1 mL of 75% ethanol. Ultimately, dried RNA-pellet was solved in diethyl phosphorocyanide treated water. We characterized RNA concentration of refined with optical density, adjusted at 260 and 280 nm wavelengths.
3.3. cDNA Synthesis
We synthesized cDNA using RevertAidTM First Strand cDNA synthesis kit (MBI, Fermentas, Lithuania). In summary, we treated RNA using DNase I (Invitrogen, Carlsbad, CA, USA) ere cDNA synthesis occurred to eschew DNA pollution and retrotranscripted cDNA in 20 µL reaction solution while it contained 5 µg total RNA, reaction buffer, RNase inhibitor (20 unit), dNTP mix (20 nM), random hexamer primer, oligo (dt) primer, and 200 unit M-MuLV reverse transcriptase. We performed reverse transcription (RT) at 42°C during 60 minutes and was terminated by raising the temperature to 70°C for 5 minutes.
3.4. Quantitative Real Time PCR
The free online Primer BLAST software (fast prim6) was used for designing particular primer sequences P53 and Hdm2 and β-actin ribosomal RNA, and gene confirmation stage was augmented using GeneDetect® oligonucleotide gene probes (Oligo 3) were prepared in triplicate as following: Hdm2 primers: 5’- CAGAGCCAAGCGGCGGCAGA-3’ (forward primer) and 5’- AGAAGCTGCTGGTGGCGGGG-3’ (reverse primer), P53 primers: 5’- TGGGCGTGAGCGCTTCGAGA-3’ (forward primer) and 5’- GGTGGCTGG AGTGAGCCCTGC-3’ (reverse primer), β-actin primers: 5’- TCCCTGGAGAAGAGCTACG-3’ (forward primer) and 5’- GTAGTTTCGTGGATGCCACA-3’ (reverse primer). For an internal control Beta-actin was used rRNA and determined the dependent gene expression using the 2-ΔCtformula:
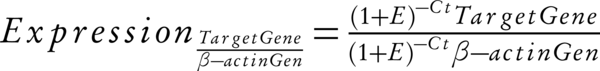
All reactions of the Real-time PCR were conducted in reaction tubes filled with 2x SYBER GREEN PCR master mix reagent (ABI, Vernon, CA, USA), 190 nM primer and 1 µg cDNA in 20 µL reaction volume. Quantitative RT PCR was carried out by Corbett Rotor-Gene 6000 thermal cycler (Corbett life science, Sydney, Australia). Then thermal cycle performed, the first cycle was at 95°C for 5 minutes, then 42 cycles of 95°C for 20 seconds (denaturation), 60°C for 20 seconds (annealing), and 72°C for 20 seconds (extension). After each test, using melting curve analysis with Corbett rotor-gene 6000 software accuracy of reaction was confirmed (Corbett Life Science, Sydney, Australia).
3.5. Irradiation
Having finished plating of the Cells (1 × 105/well) into 6-well culture plates, we incubated them for 16 hours at 37°C. Then the cells were irradiated (0, 0.5, 1, 2, 4, or 6 Gy in a single fraction) with 60-Co γ-rays beams (Clinac 21EX; Varian, Palo Alto, CA) whose dose rate was 89.514 cGy/min. When the irradiation continue to the 3rd day, to determine the viable cells, the cells triplet cultures for each combination treatment were counted.
3.6. MTT Assay
We seeded all 8 × 103 cells in each well of 96-well plate and permitted to attach during next 24 hours. Then cells were irradiated for another 24 hours and the rate of the cellular proliferation was evaluated during 24 hours (16). After adding 10 μL of 5 mg/mL MTT (3-(4, 5-dimetylthiazol-2-yl)-2, 5-diphenyltrazolium bromide) (Sigma-Aldrich,St. Louis, MO, US) to each well and incubating them at 5% CO2 and 37°C for 4 hours and after discarding the media, 200 µL of dimethyl sulfoxide appended to each and every well to solubilize the colored formazan product. 25 µL Sorenson buffer was appended to each well as solubilizer buffer. Ultimately, we got the absorbance rate with an ELISA plate reader (BioTeck, Bad Friedrichshall, Germany) at 570 nm wavelength. We compared all the computed data with the untreated cells then analyzed and normalized them.
3.7. Clonogenic Cell Survival Assay
We used clonogenic survival assay for investigation the radiosensitivity of the TE1, TE8 and TE11 cell lines. Next we plated the cells in six-well plates at an appropriate cell density. After that in the next 24 hours attached and by 60-Co γ-rays beams exposed subsequently (Clinac 21EX; Varian, Palo Alto, CA) with dose rate of 89.514 cGy/min (0, 0.5, 1, 2, 4, or 6 Gy in a single fraction). After that, we washed the wells and added new media. We then incubated renewed culture in a humidified incubator for the next two weeks (at 37°C). We enumerated the generated colonies since we fixed them with ethanol and colored with 2% Giemsa. Each of them contained more than 50 cells were enumerated as survivors. Then survival fraction and plating efficiency were calculated. Sensitizer increase proportion was specified according to the required radiation does for taking a particular surviving fraction (SF). All data points were acquired in triplicates. We normalized the gained data using the sham treated control cell plating efficiencies and applied the plot survival curves using the GraphPad Prism 6 (GraphPad Software, Inc., San Diego, CA, USA).
3.8. Statistical Analysis
Statistical analyses were performed by two-way separated pair two-tailed test and ANOVA, by Prism (version 4.0) from graph Pad. Data were explicated as mean ± SE. P < 0.05 were considered significant.
4. Results
By comparing Hdm2 gene expression between the three cell lines Te1, Te8, and Te11, significant reduction in the level of gene expression Hdm2, increased cell death with cobalt-60 gamma radiation, and an increase in the radiation sensitivity compared with the other group were observed.
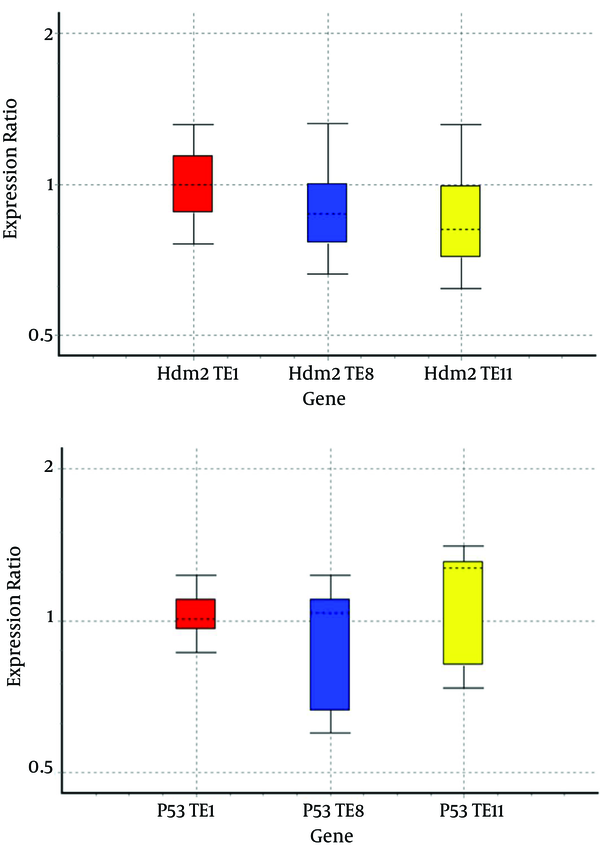
We analyzed the expression profiles of mRNA of Hdm2 and P53 in cell lines by real-time PCR assay and relative gene expressions were calculated by normalizing the results with β-actin mRNA expression (Figure 1).
mRNA Expression Levels of Hdm2 and P53 by Real Time PCR Assay
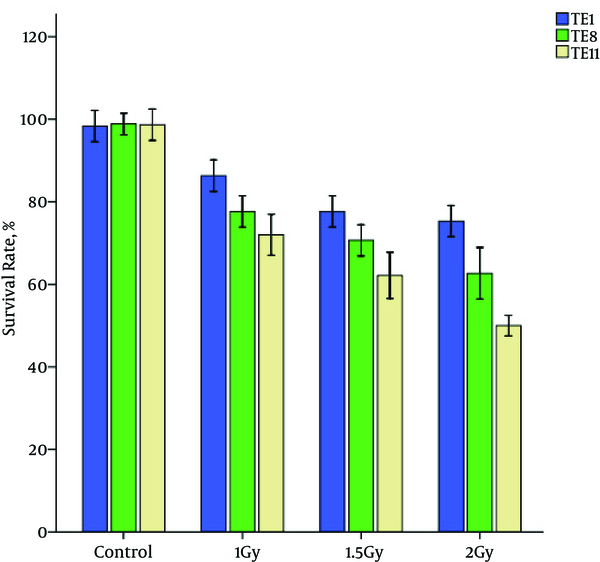
Cytotoxicity assays showed decrease in cell viability in the TE1, TE8 and TE11 cell lines with an increase in radiation dose. The following figure shows the survival rate of esophageal cancer cell lines (Figure 2).
The Viability Rate of TE1, TE8 and TE11 Cell Lines in Radiation and Control Groups
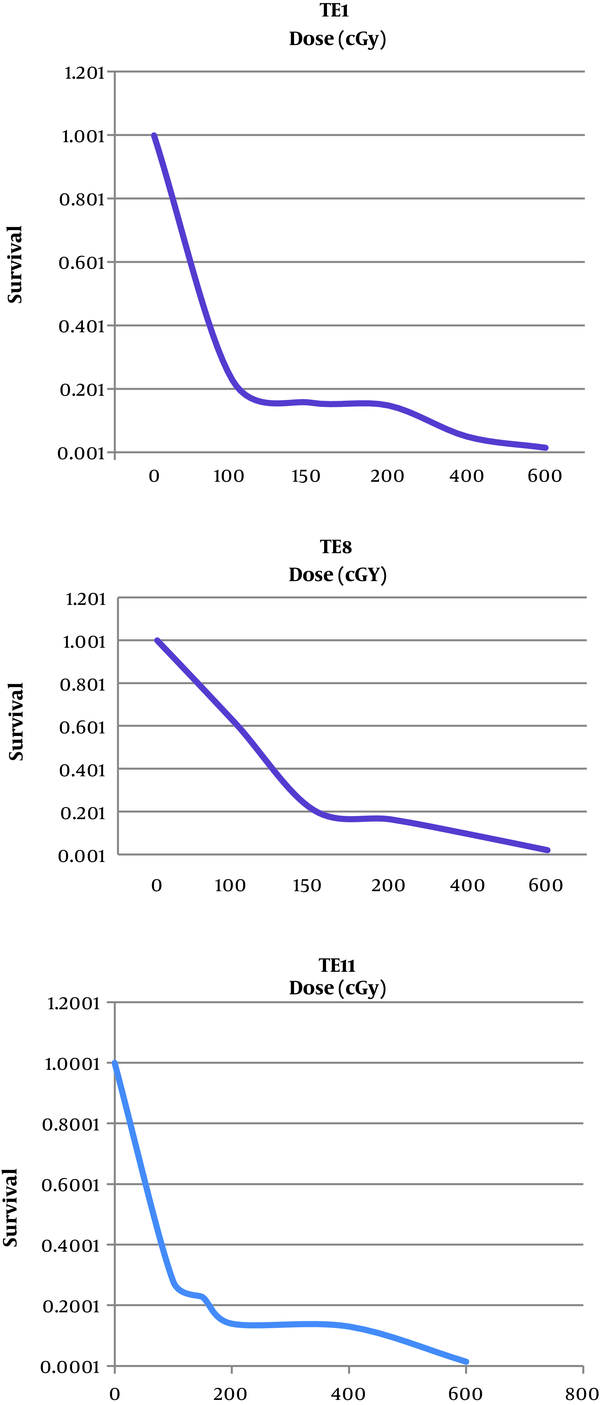
Radio-sensitivity of TE1, TE8 and TE8 were studied by a colony formation assay. We displayed the whole cell lines radiation dose-effect in Figure 3, and showed the average survival curve parameters in Figure 3.
The Effects of Radiation on Survival Rate of TE1, TE8 and TE11 Cell Lines
5. Discussion
Since Hdm2 gene is an inhibitor for P53 and with its expression it can stimulate cell proliferation, cell survival and increase the formation of tumors and from there the three cell lines TE1, TE8 and TE11 has a different expression levels of genes Hdm2; therefore, among the three cell lines decreasing the expression of the gene cause the increased radiosensitivity.
Esophageal squamous cell carcinomas have diverse radiosensitivity. Also there exists a broad variation in the esophageal cancer’s radiosensitivity of the analogous histological type. Radioresistanceacts is an obstacle in treatment of esophageal cancer, and it affects the curability of patients in neoadjuvant settings. Hence, it is vital to clarify the radio resistance mechanisms to make patients who suffere from prognosis of esophageal cancer better (8). The response of cells to the radiation is interceded by genes that manage complex regulatory pathways. All of these pathways can include cell cycle detention, DNA and apoptosis damage, and repair it since radiation (17, 18). Induced changes from radiation in expression levels of many genes and cellular RNA have been observed (9). Several genes were involved in operation of response mechanism of eukaryotic cells to IR. The mentioned genes subtend a quantity of cell cycle, checkpoint, and DNA repair genes, further mediators of apoptosis, like p53, bax, and Bcl-2. The repressions or expressions of relevant genes are dependent on cell death or cell survival in simple model systems; however, pour no light on intercellular incidents in in-situ tumors or in the clinical results of radiotherapy (1, 2, 19, 20).
Hdm2 is a vital part of the responds to radiation of type of UV and ionizing. The reduced levels of Hdm2 sensitize the cells on IR. Therefore, Hdm2 is a possible aim for therapeutic intermediation since the inhibition can increase radiosensity the subset of tumors expressing wild type of human (wt) p53, making radiotherapy more effective (2, 21). For recognizing involved genes in radiosensitivity and getting more effective radiotherapy in esophageal cancer, the radiosensitivities of 3 human esophageal cancer cell lines were studied. TE1, TE8 and TE11 with Hdm2 gene expression levels are different, so it can be an appropriate model based on molecular radiobiology techniques in the perusal gene Hdm2 appeasing and improving the radiation sensitivity of the cells gained. Due to increased Hdm2 expression, this gene in these cells can be used as a cellular marker. Ogawa et al. classified esophageal cancer cells to 13 categories (9). They found that TE11 cells are more sensitive to radiation than other cell lines. From these cell lines, TE-11 was so sensitive to radiation. Microarray technique developments have simplified the simultaneous perusal of the expression of human genes. The outcomes of oligonucleotide microarray disclosed that at the mRNA level, 54 radiosensitivity dependent genes were up-regulated 4-fold and 17 genes were down-regulated ≥ 4-fold in TE-11 ESCC cell lines compared with other ESCC cell lines. We measured Hdm2 and P53 mRNA expression by semi-quantitative RT-PCR and then the influence expression of Hdm2 and P53 on radiosensitivity.
We studied the radiosensitivities of TE1, TE8 and TE11 cell lines, and compared the changes of Hdm2 gene expression. The results indicated decrease in Hdm2 gene expression in TE11 and increase in P53 gene expression lead to cell radiosensitization that was probably dependent on its effects on cell apoptosis as explaned in the previous section. Furthermore, TE1 and TE8 up-regulated of Hdm2 and down-regulated of P53 lead to cell radioresistance. After 24 hours radiation, MTT assay showed that the cell viability had different degrees decrease in the TE1, TE8 and TE11 cells, while the viability of TE11 had change compared with the control group. The overexpression of Hdm2 can decrese apoptosis in Esophageal squamous cell carcinomas. Gained data illustrated that the Hdm2 overexpression almost decresed the apoptotic rate of Esophageal squamous cell carcinomas in response to radiation; whereas, suppression of P53 transcriptional function mediates poor therapeutic response to radiation. A previous study has demonstrated that APE1 expression has been found to be associated with radioresistance in tumors (22) and mi-34a is a DNA damage-responsive gene. IR influenced its expression. Kato and colleagues disclosed that Caenorhabditis elegans with loss-of-function mutations in the mir-34 gene have odd cellular survival responses to radiation. Those animals are greatly radiosensitive in the soma and radioresistant in the gremlin (23). Previous data confirmed that the effect of hdm2 decrease on multitude p53-dependent and apoptosis-related genes remain obscure, as the does mechanism of hdm2-siRNA-induced apoptosis in human cancers (24). In this paper we illustrated that the response of cells to IR depends on the IR dose and Hdm2 expression level. High expression continue transcriptional operation for repairing the DNA damage since post-irradiation and a little expression would induce apoptosis directly.
5.1. Conclusion
Hdm2 protein binding to the P53 protein is a key regulator of cell cycle control and genome protection cell cycle control, programmed cell death and response cells to radiation play important roles. But moiety of the human tumors express wild type (wt) p53, and its activation by antagonizing its negative regulator Hdm2 potency propose a new therapeutic method (25). Our experiments displayed that the possibility of this view point to vitro, while extension of pharmacological inhibitors again is a challenge.
Acknowledgements
References
-
1.
Pirayesh Islamian J, Mohammadi M, Baradaran B, Farajollahi A, Aghamiri SM, Asghari Jafarabadi M, et al. Enhancing radiosensitivity of TE1, TE8, and TE 11 esophageal squamous carcinoma cell lines by Hdm2-siRNA targeted gene therapy in vitro. Bioimpacts. 2016;6(2):93-8. [PubMed ID: 27525226]. https://doi.org/10.15171/bi.2016.13.
-
2.
Islamian JP, Mohammadi M, Baradaran B. Inhibition of human esophageal squamous cell carcinomas by targeted silencing of tumor enhancer genes: an overview. Cancer Biol Med. 2014;11(2):78-85. [PubMed ID: 25009749]. https://doi.org/10.7497/j.issn.2095-3941.2014.02.002.
-
3.
Poynton AR, Walsh TN, Kelly A, Harney M, Stuart R, Daly PA. Small cell carcinoma of the oesophagus. Euro J Surgical Oncol. 1997;23(6):509-12.
-
4.
Hagiwara N, Tajiri T, Tajiri T, Miyashita M, Sasajima K, Makino H, et al. Biological behavior of mucoepidermoid carcinoma of the esophagus. J Nippon Med Sch. 2003;70(5):401-7. [PubMed ID: 14578940].
-
5.
Bosset JF, Lorchel F, Mantion G. Neoadjuvant treatment of early stage squamous cell carcinoma of the esophagus. Dis Esophagus. 2002;15(2):117-20. [PubMed ID: 12220417].
-
6.
Xiao ZF, Yang ZY, Liang J, Miao YJ, Wang M, Yin WB, et al. Value of radiotherapy after radical surgery for esophageal carcinoma: a report of 495 patients. Ann Thorac Surg. 2003;75(2):331-6. [PubMed ID: 12607634].
-
7.
Medical Research Council Oesophageal Cancer Working G. Surgical resection with or without preoperative chemotherapy in oesophageal cancer: a randomised controlled trial. Lancet. 2002;359(9319):1727-33. [PubMed ID: 12049861]. https://doi.org/10.1016/S0140-6736(02)08651-8.
-
8.
Herskovic A, Martz K, al-Sarraf M, Leichman L, Brindle J, Vaitkevicius V, et al. Combined chemotherapy and radiotherapy compared with radiotherapy alone in patients with cancer of the esophagus. N Engl J Med. 1992;326(24):1593-8. [PubMed ID: 1584260]. https://doi.org/10.1056/NEJM199206113262403.
-
9.
Ogawa R, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Mori Y, et al. Identification of candidate genes involved in the radiosensitivity of esophageal cancer cells by microarray analysis. Dis Esophagus. 2008;21(4):288-97. [PubMed ID: 18477249]. https://doi.org/10.1111/j.1442-2050.2007.00759.x.
-
10.
Lam AK. Molecular biology of esophageal squamous cell carcinoma. Crit Rev Oncol Hematol. 2000;33(2):71-90. [PubMed ID: 10737369].
-
11.
Mandard AM, Hainaut P, Hollstein M. Genetic steps in the development of squamous cell carcinoma of the esophagus. Mutat Res. 2000;462(2-3):335-42. [PubMed ID: 10767643].
-
12.
Futami T, Miyagishi M, Seki M, Taira K. Induction of apoptosis in HeLa cells with siRNA expression vector targeted against bcl-2. Nucleic Acids Res Suppl. 2002;(2):251-2. [PubMed ID: 12903200].
-
13.
Nejati-Koshki K, Akbarzadeh A, Pourhasan-Moghaddam M, Abhari A, Dariushnejad H. Inhibition of leptin and leptin receptor gene expression by silibinin-curcumin combination. Asian Pac J Cancer Prev. 2014;14(11):6595-9. [PubMed ID: 24377502].
-
14.
Nasiri M, Zarghami N, Koshki KN, Mollazadeh M, Moghaddam MP, Yamchi MR, et al. Curcumin and silibinin inhibit telomerase expression in T47D human breast cancer cells. Asian Pac J Cancer Prev. 2013;14(6):3449-53. [PubMed ID: 23886126].
-
15.
Yuan Y, Liao YM, Hsueh CT, Mirshahidi HR. Novel targeted therapeutics: inhibitors of MDM2, ALK and PARP. J Hematol Oncol. 2011;4:16. [PubMed ID: 21504625]. https://doi.org/10.1186/1756-8722-4-16.
-
16.
Aghaee F, Pirayesh Islamian J, Baradaran B. Enhanced radiosensitivity and chemosensitivity of breast cancer cells by 2-deoxy-d-glucose in combination therapy. J Breast Cancer. 2012;15(2):141-7. [PubMed ID: 22807930]. https://doi.org/10.4048/jbc.2012.15.2.141.
-
17.
Teyssier F, Bay JO, Dionet C, Verrelle P. [Cell cycle regulation after exposure to ionizing radiation]. Bull Cancer. 1999;86(4):345-57. [PubMed ID: 10341340].
-
18.
Fekri Aval S, Akbarzadeh A, Yamchi MR, Zarghami F, Nejati-Koshki K, Zarghami N. Gene silencing effect of SiRNA-magnetic modified with biodegradable copolymer nanoparticles on hTERT gene expression in lung cancer cell line. Artif Cells Nanomed Biotechnol. 2016;44(1):188-93. [PubMed ID: 25020049]. https://doi.org/10.3109/21691401.2014.934456.
-
19.
Wang XC, Wang W, Zhang ZB, Zhao J, Tan XG, Luo JC. Overexpression of miRNA-21 promotes radiation-resistance of non-small cell lung cancer. Radiat Oncol. 2013;8:146. [PubMed ID: 23777591]. https://doi.org/10.1186/1748-717X-8-146.
-
20.
Zonouzi AAP, Nejatizadeh A, Rahmati-Yamchi M, Fardmanesh H, Shakerizadeh S, Zonouzi AP. Dysregulated expression of Dicer in invasive ductal breast carcinoma. Med Oncol. 2015;32(7):1-7.
-
21.
Perry ME. Mdm2 in the response to radiation. Mol Cancer Res. 2004;2(1):9-19. [PubMed ID: 14757841].
-
22.
Cun Y, Dai N, Xiong C, Li M, Sui J, Qian C, et al. Silencing of APE1 enhances sensitivity of human hepatocellular carcinoma cells to radiotherapy in vitro and in a xenograft model. PLoS One. 2013;8(2):55313. [PubMed ID: 23418439]. https://doi.org/10.1371/journal.pone.0055313.
-
23.
Duan W, Xu Y, Dong Y, Cao L, Tong J, Zhou X. Ectopic expression of miR-34a enhances radiosensitivity of non-small cell lung cancer cells, partly by suppressing the LyGDI signaling pathway. J Radiat Res. 2013;54(4):611-9. [PubMed ID: 23349340]. https://doi.org/10.1093/jrr/rrs136.
-
24.
Liu TG, Yin JQ, Shang BY, Min Z, He HW, Jiang JM, et al. Silencing of hdm2 oncogene by siRNA inhibits p53-dependent human breast cancer. Cancer Gene Ther. 2004;11(11):748-56. [PubMed ID: 15375377]. https://doi.org/10.1038/sj.cgt.7700753.
-
25.
Ozaki T, Nakagawara A. the attractive tumor suppressor in the cancer research field. BioMed Res Inter. 2010:53.