High-coverage genomes of all extant penguin taxa
Dataset type: Genomic
Data released on September 11, 2019
Pan H; Cole T; Bi X; Fang M; Zhou C; Yang Z; Ksepka DT; Hart T; Bouzat JL; Argilla LS; Bertelsen MF; Boersma PD; Bost C; Cherel Y; Dann P; Fiddaman SR; Howard P; Labuschagne K; Mattern T; Miller G; Parker P; Phillips RA; Quillfeldt P; Ryan PG; Taylor H; Thompson DR; Young MJ; Ellegaard MR; Gilbert MTP; Sinding MS; Pacheco G; Shepherd LD; D Tennyson AJ; Grosser S; Kay E; Nupen LJ; Ellenberg U; Houston DM; Reeve AH; Johnson K; Masello JF; Stracke T; McKinlay B; García Borboroglu P; Zhang DX; Zhang G (2019): High-coverage genomes of all extant penguin taxa GigaScience Database. https://doi.org/10.5524/100649
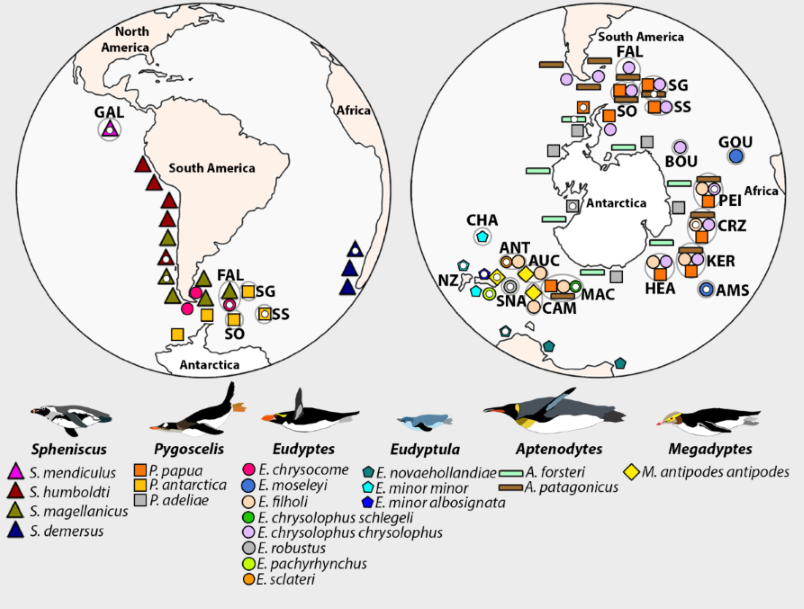
Penguins (Sphenisciformes) are a highly diverse order of seabirds distributed widely across the Southern Hemisphere. They shared a common ancestor with Procellariiformes about 60 million years ago, and have subsequently transited from flying seabirds to flightless marine divers. Approximately 20 extant penguin species are recognized across six well-defined genera, ranging from the Galápagos Islands on the equator, to the oceanic temperate forests of New Zealand, the rocky coastlines of the sub-Antarctic islands, and the sea-ice around Antarctica. To inhabit such diverse and extreme environments, this speciose order has evolved many physiological and morphological adaptations. However, penguins are also highly sensitive to climate change, and most species are already declining or are predicted to decline under future climate change scenarios. Therefore, penguins are an exciting system for understanding the evolutionary processes of speciation, adaptation and demography. Genomic data are an emerging resource for addressing questions about such processes. Here we present 19 new high-coverage genomes that, together with two previously published genomes also available in GigaDB, encompass all extant penguin species. As such, this dataset provides a novel resource for understanding the evolutionary history within penguins, and between penguins and other avifauna. We believe that our dataset and project will be important for cultural heritage and the conservation of this iconic Southern Hemisphere species assemblage. Comparative and evolutionary genomic analyses are currently being carried out, and the consortium welcomes new members interested in contributing to this work. While this work is still underway we have published these 19 penguin genomes to provide early-release, while requesting researchers intending to use this data for similar cross-species comparisons to continue to follow the Fort Lauderdale and Toronto rules.
Keywords:
Additional details
Read the peer-reviewed publication(s):
- Pan, H., Cole, T. L., Bi, X., Fang, M., Zhou, C., Yang, Z., Ksepka, D. T., Hart, T., Bouzat, J. L., Argilla, L. S., Bertelsen, M. F., Boersma, P. D., Bost, C.-A., Cherel, Y., Dann, P., Fiddaman, S. R., Howard, P., Labuschagne, K., Mattern, T., … Zhang, G. (2019). High-coverage genomes to elucidate the evolution of penguins. GigaScience, 8(9). https://doi.org/10.1093/gigascience/giz117 (PubMed:31531675)
Related datasets:
doi:10.5524/100649 Cites doi:10.5524/100006
doi:10.5524/100649 Cites doi:10.5524/100005
doi:10.5524/100649 HasPart doi:10.5524/102164
doi:10.5524/100649 HasPart doi:10.5524/102165
doi:10.5524/100649 HasPart doi:10.5524/102166
doi:10.5524/100649 HasPart doi:10.5524/102167
doi:10.5524/100649 HasPart doi:10.5524/102168
doi:10.5524/100649 HasPart doi:10.5524/102169
doi:10.5524/100649 HasPart doi:10.5524/102170
doi:10.5524/100649 HasPart doi:10.5524/102171
doi:10.5524/100649 HasPart doi:10.5524/102172
doi:10.5524/100649 HasPart doi:10.5524/102173
doi:10.5524/100649 HasPart doi:10.5524/102174
doi:10.5524/100649 HasPart doi:10.5524/102175
doi:10.5524/100649 HasPart doi:10.5524/102176
doi:10.5524/100649 HasPart doi:10.5524/102177
doi:10.5524/100649 HasPart doi:10.5524/102178
doi:10.5524/100649 HasPart doi:10.5524/102179
doi:10.5524/100649 HasPart doi:10.5524/102180
doi:10.5524/100649 HasPart doi:10.5524/102181
doi:10.5524/100649 HasPart doi:10.5524/102182
Click on a table column to sort the results.
Table Settings| File Name | Description | Sample ID | Data Type | File Format | Size | Release Date | File Attributes | Download |
|---|---|---|---|---|---|---|---|---|
| Readme | TEXT | 4.17 kB | 2019-08-27 | MD5 checksum: 7156bfdafb3307f067f80b44f26e2236 |
||||
| phylogenetic tree file of all species of penguins, in newick format | Phylogenetic tree | Newick | 1.82 kB | 2019-09-17 | MD5 checksum: 798a83a888a4d1cbf1ef19ddb34c7e1d |
|||
| alignment file of all common orthologous genes found in all species of penguins. Used to generate the phylogenetic tree. | Alignments | UNKNOWN | 11.32 MB | 2019-09-17 | MD5 checksum: 0069b6b648c970b93de7c6a811a37d55 |
| Funding body | Awardee | Award ID | Comments |
|---|---|---|---|
| Carlsbergfondet | G Zhang | CF CF16-0663 | |
| Ministry of Science and Technology (MOST) | G Zhang | 2018YFC1406901 | National Key Research and Development Program |
| Chinese Academy of Sciences | G Zhang | XDB13000000 | Strategic Priority Research Program |
| Chinese Academy of Sciences | G Zhang | XDB31020000 | Strategic Priority Research Program |
| Lundbeckfonden | G Zhang | R190-2014-2827 | |
| Villum Fonden | G Zhang | 25900 | |
| European Research Council | MTP Gilbert | 681396 | Extinction Genomics - Consolidator Grant |
| Date | Action |
|---|---|
| September 11, 2019 | Dataset publish |
| September 17, 2019 | Additional file MPEST.nwk added |
| September 17, 2019 | Additional file mafft_concat_No_Missing_Data.phylip.gz added |
| September 17, 2019 | mafft_concat_No_Missing_Data.phylip.gz: additional file attribute added |
| September 17, 2019 | File mafft_concat_No_Missing_Data.phylip.gz updated |
| September 17, 2019 | MPEST.nwk: additional file attribute added |
| September 17, 2019 | File MPEST.nwk updated |
| October 14, 2022 | Manuscript Link updated : 10.1093/gigascience/giz117 |