Supporting data for "Chiron: Translating nanopore raw signal directly into nucleotide sequence using deep learning"
Dataset type: Software
Data released on April 10, 2018
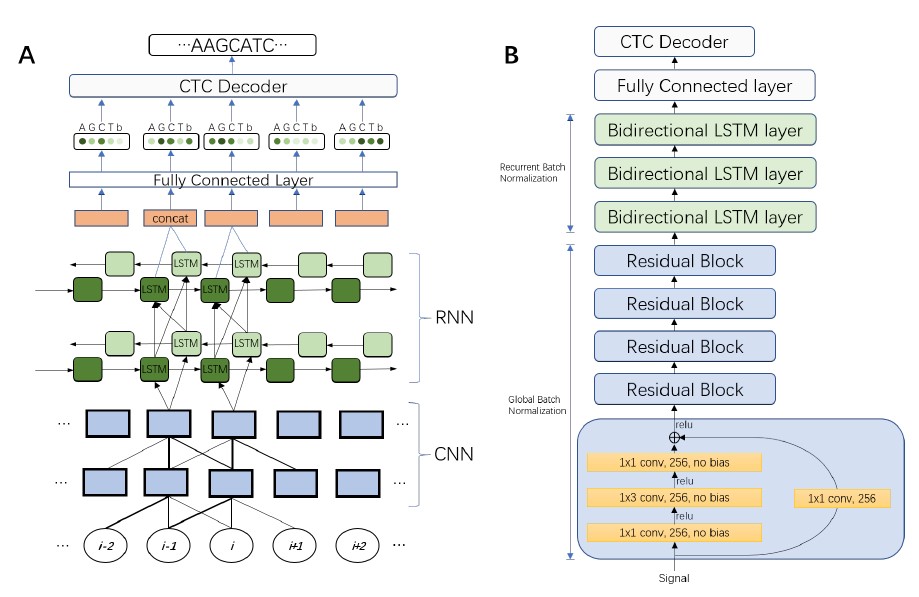
Sequencing by translocating DNA fragments through an array of nanopores is a rapidly maturing technology which offers faster and cheaper sequencing than other approaches. However, accurately deciphering the DNA sequence from the noisy and complex electrical signal is challenging. Here, we report Chiron, the first deep learning model to achieve end-to-end basecalling: directly translating the raw signal to DNA sequence without the error-prone segmentation step. Trained with only a small set of 4000 reads, we show that our model provides state-of-the-art basecalling accuracy even on previously unseen species. Chiron achieves basecalling speeds of over 2000 bases per second using desktop computer graphics processing units, making it competitive with other deep-learning basecalling algorithms.
Additional details
Read the peer-reviewed publication(s):
- Teng, H., Cao, M. D., Hall, M. B., Duarte, T., Wang, S., & Coin, L. J. M. (2018). Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. GigaScience, 7(5). https://doi.org/10.1093/gigascience/giy037 (PubMed:29648610)
- Teng, H., Cao, M. D., Hall, M. B., Duarte, T., Wang, S., & Coin, L. J. M. (2019). Correction to: Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. GigaScience, 8(5). https://doi.org/10.1093/gigascience/giz049 (PubMed:31077312)
Additional information:
https://github.com/haotianteng/chiron
https://pypi.python.org/pypi/chiron
https://github.com/nanopore-wgs-consortium/NA12878
Accessions (data included in GigaDB):
BioProject: PRJNA386696
SRA: SRP136964
Click on a table column to sort the results.
Table Settings| Sample ID | Common Name | Scientific Name | Sample Attributes | Taxonomic ID | Genbank Name |
|---|---|---|---|---|---|
| MT20823 | Mycobacterium tuberculosis | Geographic location (longitude):143.12 Geographic location (latitude):-9.05 Geographic location (country and/or sea,region):Pa... ... |
1773 | ||
| NA12878 | Human | Homo sapiens | Description: Age:not provided Source material identifiers:Coriell:NA12878 ... |
9606 | human |
Click on a table column to sort the results.
Table Settings| File Name | Description | Sample ID | Data Type | File Format | Size | Release Date | File Attributes | Download |
|---|---|---|---|---|---|---|---|---|
| Readme | TEXT | 2.09 kB | 2018-03-08 | MD5 checksum: dca652223f64af8b740b221113fd84eb |
||||
| Evaluation dataset of E.coli and Lambda Phage, files is in same format as the train dataset. | Mixed archive | TAR | 480.05 MB | 2018-03-08 | MD5 checksum: 908bb5c318d36b4c53b367f50b7e8942 |
|||
| Training dataset of E.coli and Lambda Phage | Mixed archive | TAR | 469.46 MB | 2018-03-08 | MD5 checksum: 76fe29b398d735cfee7b6cd98b282d3a |
|||
| Archival copy of the GitHub repository https://github.com/haotianteng/chiron downloaded 7-March-2018.. A basecaller for Oxford Nanopore Technologies' sequencers. | GitHub archive | archive | 85.51 MB | 2018-03-08 | RRID: SCR_015950 MD5 checksum: 4d60da15a8feb2e826ed74b9f4331a1f |
|||
| Archival copy of the GitHub repository https://github.com/nanopore-wgs-consortium/NA12878 downloaded 7-March-2018. Oxford Nanopore Human Reference Datasets, data hosted on AWS. License of these data are CC-BY4 | GitHub archive | archive | 2.71 MB | 2018-03-08 | license: CC-BY4.0 MD5 checksum: 60d89200f802cd4d9ce48e74a186a363 |
|||
| This is the benchmark dataset of assembly identity rate and relative length ratio among basecallers | Mixed archive | GZIP | 544.72 MB | 2018-03-19 | MD5 checksum: 4cefe62c7d4ddd761524bbca8b1ef2f3 |
|||
| This is the benchmark dataset of read accuracy accross different basecallers among 4 species. | Mixed archive | GZIP | 26.53 GB | 2018-03-19 | MD5 checksum: aecbc5a6e639d7173acdebf509711433 |
| Funding body | Awardee | Award ID | Comments |
|---|---|---|---|
| National Health and Medical Research Council | LJM Coin | GNT1130084 | |
| Australian Research Council | LJM Coin | DP170102626 | |
| MB Hall | Westpac Future Leaders Scholarship |
| Date | Action |
|---|---|
| April 10, 2018 | Dataset publish |
| July 4, 2018 | Manuscript Link added : 10.1093/gigascience/giy037 |
| August 31, 2019 | Manuscript Link added : 10.1093/gigascience/giz049 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| August 5, 2020 | Sample Attribute added : of Sample MT20823 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| September 24, 2021 | Sample Attribute added : of Sample NA12878 |
| October 14, 2022 | Manuscript Link updated : 10.1093/gigascience/giz049 |
| November 10, 2022 | Manuscript Link updated : 10.1093/gigascience/giy037 |