Published online Sep 14, 2018. doi: 10.5306/wjco.v9.i5.90
Peer-review started: June 12, 2018
First decision: July 19, 2018
Revised: August 2, 2018
Accepted: August 6, 2018
Article in press: August 7, 2018
Published online: September 14, 2018
FMS-like tyrosine kinase 3 (FLT3) is classified as a type III receptor tyrosine kinase, which exerts a key role in regulation of normal hematopoiesis. FLT3 mutation is the most common genetic mutation in acute myeloid leukemia (AML) and represents an attractive therapeutic target. Targeted therapy with FLT3 inhibitors in AML shows modest promising results in current ongoing clinical trials suggesting the complexity of FLT3 targeting in therapeutics. Importantly, resistance to FLT3 inhibitors may explain the lack of overwhelming response and could obstruct the successful treatment for AML. Here, we summarize the molecular mechanisms of primary resistance and acquired resistance to FLT3 inhibitors and discuss the strategies to circumvent the emergency of drug resistance and to develop novel treatment intervention.
Core tip: FMS-like tyrosine kinase 3 (FLT3) mutations including internal tandem duplication (ITD) or point mutation in tyrosine kinase domain are common genetic abnormalities in acute myeloid leukemia (AML), predicting dismal outcome. The Federal Drug Administration granted the use of Midostaurin (Novartis) in newly diagnosed FLT3-ITD positive AML in April 2017. A number of other FLT3 inhibitors are in different phases of clinical trials. However, emerging drug resistance poses a major challenge for clinicians to use FLT3 inhibitors. In this manuscript, we systematically reviewed mechanism of primary resistance and acquired resistance to FLT3 inhibitors. We then propose different strategies to overcome drug resistance and novel treatment options for FLT3-ITD positive AML.
- Citation: Zhou J, Chng WJ. Resistance to FLT3 inhibitors in acute myeloid leukemia: Molecular mechanisms and resensitizing strategies. World J Clin Oncol 2018; 9(5): 90-97
- URL: https://www.wjgnet.com/2218-4333/full/v9/i5/90.htm
- DOI: https://dx.doi.org/10.5306/wjco.v9.i5.90
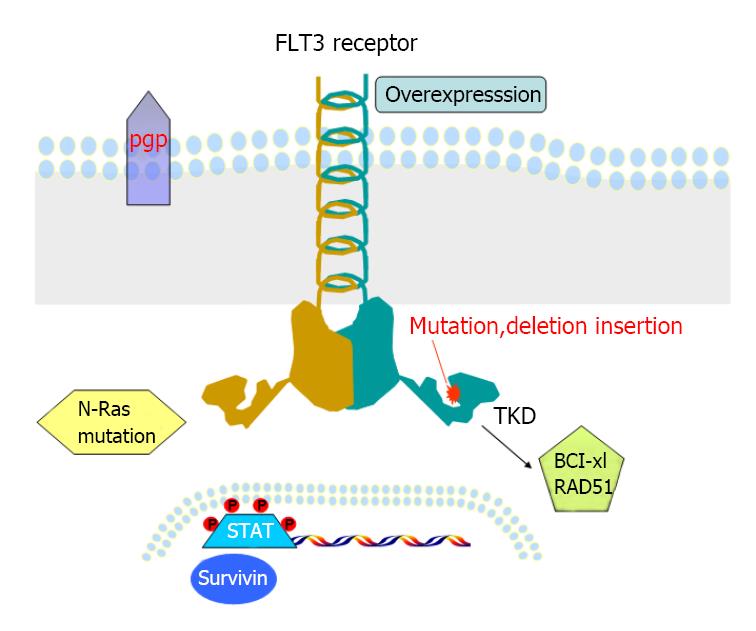
Acute myeloid leukemia (AML) consists of a group of different disease characterized with diverse cellular morphologies and various genetic abnormalities[1-5]. Many of these genetic lesions are of clinical importance because they not only implicate in the pathology of AML, but also have prognostic values[6-8]. Mutation in in FMS-like tyrosine kinase 3 (FLT3) confers inferior response to chemotherapy and poor overall survival in AML patients[9-11]. Since the discovery of FLT3 mutations in 1996[12], intensive research effort has provided a better understanding of the molecular mechanism of normal and aberrant FLT3 signaling transduction pathways. Internal tandem duplications (ITDs) in the juxtamembrane domain and activating point mutations in the second tyrosine kinase domain (TKD) occur in near 30% and 10% of patients with AML respectively[13-15].
FLT3 mutations constitutively activate PI3K-AKT, RAS-MEK-MAPK, and STAT5 pathways and result in uncontrolled cell proliferation and cell survival[16-19]. On the other hand, FLT3 mutations suppress myeloid transcription factors PU.1, CCAAT/enhancer-binding protein α (C/EBPα), which result in blocking of myeloid differentiation[20,21]. Thus, FLT3 mutations exert a key role in the pathology of AML, and have been validated as promising intervening targets[22-25]. Currently, Midostaurin (PKC412, Novartis) has been granted by the Federal Drug Administration (FDA) in the use in newly diagnosed FLT3-ITD positive AML in combination with chemotherapy[26]. Moreover, there are about a dozen of other FLT3 inhibitors in different phases of clinical development[27]. Despite most FLT3 inhibitors display strong effectiveness in cell culture system, most of AML patients in trials haven’t achieved durable response[28-30]. Notably, AML patients inevitably don’t respond to these drugs when they are administrated as single agent for a period. Scientists first observed this resistance phenomenon in patients with chronic myeloid leukemia (CML) who received imatinib mesylate (Gleevec), the first small molecule kinase inhibitor targeting BCR-ABL fusion protein[31].
Here we review published literature on preclinical and clinical findings and molecular mechanisms of primary resistance and acquired resistance to FLT3 inhibitors. We further discuss the strategies to circumvent the emergency of drug resistance and development of novel treatment intervention.
The identification of a number of de novo and secondary point mutations in the BCR-ABL kinase domain from imatinib-resistant patients promotes researchers to investigate variable sensitivity of FLT3 inhibitors between different activating point mutations in the kinase domain of FLT3.
Based on the mutations identified in AML cases, Grundler et al[32] employed site-directed mutagenesis method to create Asp835Tyr, Ile836del and Ile836Met + Arg (numbering is based on the human FLT3) into the cDNA of murine wild-type FLT3. These vectors were then transfected into murine Ba/F3 and 32Dcl3 cells, rendering them independent from growth factors. Tyrphostin AG1296 does not inhibit FLT3 Asp835Tyr (D835Y)-induced proliferation, inhibition of apoptosis, as well as the downstream signaling, and phosphorylation of STAT5. AG1296 is effective on the inhibition of signaling from FLT3 Ile836del (I836del), -ITD and to the less extent, from Ile836Met + Arg (I836M + R). Staurosporin derivative PKC412 is sensitive to all the 3 catalytic domain mutations, but less sensitivity to - ITD mutant. Indolinone compound SU5416 shows similar inhibition profile as PKC412. This study suggests that different inhibitors exert a divergent sensitivity toward different mutations in the FLT3 receptors.
A similar approach was used to introduce each FLT3 activation loop mutant, including D835Y, Asp835Ala (D835A), Asp835Glu (D835E), Asp835Gly (D835G), Asp835His (D835H), Asp835Asn (D835N), Asp835Val (D835V), and D835del into human FLT3 cDNA[33]. Ba/F3 cells were transformed with each vector. These 8 activation loop mutations display variable sensitivity toward quinazoline-based inhibitor MLN518 with more than a 10-fold range. I836del is as sensitive as ITD with IC50 0.55 µmol/L. The IC50 of D835E, D835A, D835N, D835H ranges from 0.99 to 2.65 µmol/L. D835del, D835V and D835Y confer relative resistance to MLN518 with much higher IC50 up to greater than 10 µmol/L.
This phenomenon could be explained by the assumption that the mutations in the amino acid sequence change the conformation of the catalytic domain of FLT3, resulting in a weaken affinity with FLT3 inhibitors[32,33]. However, the structural analysis of these inhibitors in the context of various mutants is not available in these papers. These findings are of great clinical interesting. Patients enrolled in the FLT3 inhibitor trials potentially can be screened for all known activation loop mutations. In addition, sensitivity of specific inhibitor can potentially be evaluated ex vivo prior to clinical administration to avoid known primary resistant cases.
About 1% to 2% of newly diagnosed AML patients carry both ITD and TKD (FLT3-ITD-TKD) with worse outcome when compared with patients with either ITD or TKD mutation alone[34-36]. Similarly, an in vitro study using Ba/F3 cells transfected with FLT3-ITD-TKD dual mutants, for example ITD-D835N and ITD-D835Y, can induce resistance toward not only FLT3 inhibitor SU5614, but also cytotoxic drug Daunorubicin[37]. Molecular study reveals these dual mutants promote overactivation of STAT5 pathway, and result in upregulation of downstream target Bcl-xL and RAD51 and arrest in the G2/M phase of the cell cycle[37]. Overexpression of Bcl-2 is also detected in primary AML patient samples with FLT3-ITD-Y591 duplication, correlated to high levels of phosphorylated p53. However, whether this mutant induces resistance to FLT3 inhibitors has not been tested[38].
Other possible mechanisms of primary resistance to TKIs have been investigated. P-glycoprotein (p-gp, also named multi-drug resistance 1, MDR1), a major membrane efflux pump, Primary AML blasts co-expressing p-gp and FLT3-ITD, are resistant to herbimycin A, a tyrosine kinase inhibitor, and AG1296, but not to PKC412[39]. The difference could be due to the fact that PKC421 has dually inhibitory roles in FLT3 and protein kinase C (PKC), which can induce phosphorylation of p-gp, resulting in subversion of p-gp mediated MDR. However, other study shows no association between FLT3 mutations and high levels of MDR1 gene expression in AML patients[40].
In contrast to earlier studies, Siendones et al[41] demonstrate that inhibition of FLT3-ITD activity does not necessary block the phosphorylation of AKT, ERK and STAT5, which are the 3 major pathways activated by FLT3 mutations, in some primary AML cells. This could be one reason for the limited anti-tumor effect of FLT3 inhibitors used as monotherapy in clinical trials. In addition, a new “niche and leukemia stem cell” model was proposed to explain the limitation of single agent[42]. If FLT3 - ITD is presented in CD34 + CD38 - CD123 + leukemia stem and progenitor cells (LSPC) from primary AML samples, they are more resistant to FLT3 inhibitor in culture under defined nice conditions (fibronectin, IL - 3, SCF, IL - 6 and Ang-). This result is consistent with an earlier finding that patients whose CD34+CD33- precursors harbor FLT3 - ITD have worse outcome than patients whose CD34 + CD33 + progenitors have FLT3 - ITD[43]. These data indicate that FLT3 - ITD AML derived from the less mature progenitors may be associated with drug resistance.
Pioneer researches in imatinib-resistant CML patients revealed two different resistant mechanisms including increased copy number of BCR-ABL fusion and point mutations in its adenosine triphosphate (ATP) binding motif[44]. These initial discoveries facilitate our understanding of acquired resistance to FLT3 inhibitors. As demonstrated by imatinib-resistant CML studies, over expression of a mutated FLT3 could also be a common mechanism for drug desensitization and leading to resistance. Weisberg and Boulton et al[45] first address this issue using a Ba / F3 - FLT3 - ITD resistant polyclonal subline developed by coculture with increasing concentration of PKC412 (up to 40 nmol/L) with the parental cell line over 2 mo. The protein level of FLT3-ITD is significantly increased in this resistant subline compared to the parental Ba / F3 - FLT3 - ITD, leading to desensitize PKC412. It is not clear that FLT3 - ITD protein over expression was regulated on transcriptional (gene amplification) or post-translational levels (increased protein stability). Also, there is no further study on whether this resistant subline harbors point mutation(s) in the TKD domain.
Other resistant lines, designated as Ba / F3 - ITD - R1 to R4 derived from the same parental Ba / F3 - FLT3 - ITD have been developed in the presence of escalatory dose of SU5614[46]. The average IC50 of these lines is 17 - fold higher than the parent line. Consistent with their resistant phenotypes, on the molecular level, the phosphorylation of MAPK and STAT5 in these sublines is not inhibited by higher dose up to 10 µmol/L SU5614, while 1 µmol/L of SU5614 effectively decreases activity of MAPK and STAT5 in the parental line. They are also completely resistant to AG1295, which is structurally similar to SU5614. But, Ba / F3 - ITD - R1 to R4 display a similar sensitivity to a structural unrelated FLT3 inhibitor PKC412, a general TKI, Genistein and a chemotherapeutic agent cytosine arabinoside (Ara-C) as the parent ITD cells. Both flow cytometric analysis and western blot demonstrate elevated amount of FLT3 receptor in the resistant Ba / F3 - ITD - R1 to R4 compared with the parent line. Sequence analysis of TKD domain identifies Y842H mutation in ITD-R1 and –R2 cells, and D835 mutation in ITD-R3 and -R4 cells. These data indicate that both FLT3 target desensitization and acquired mutations in the activation kinase domain can contribute to secondary resistance in vitro.
Using random PCR mutagenesis to introduce point mutations in ATP-binding pocket of the KD of MSCV-FLT3-ITD, Cools et al[47] identified 4 different point mutations (Ala627, Asn676, Phe691, or Gly697) in Ba/F3 cells that render resistance to PKC412, SU5614 or K-252a. The G697R mutation is the most resistant clone to all the three inhibitors tested. Accordingly, PCK412 fails to reduce phosphorylation of FLT3 up to 400 nmol/L in cells with G697R and they are cross-resistant to other structurally different FLT3 inhibitors (GTP-14546, AGL2043, D-64406, D-64476, TMPPP and DQPPC). Modeling crystal structure of FLT3 receptor in complex with PCK412 indicates that the amino acid Gly697 and Phe691 directly contact with PKC412 and substitution Gly697 with a larger amino acid will decrease its binding affinity due to possible steric clash with the FLT3 inhibitors. Importantly, mutation in Asn676 (N676K) has been reported in 1 of 6 patients with FLT3-ITD AML who relapsed after PKC412 treatment in a phase 2 clinical trial[48]. The authors were able to rule out other common mechanisms of drug resistance including gene amplification, overexpression of FLT3 protein, drug metabolism, drug efflux, inhibition by serum proteins and major deficiency in apoptosis pathway. Although the identification of N676K is clinically significant to elucidate the mechanism of resistance and relapse, so far acquired point mutation of TKD has been reported only in a FLT3 inhibitor treated, and relapsed AML patient. In addition, transfection of FLT3-ITD-N676K in 32D cells confers resistance to PKC412[48]. This finding is in consistent with the clinical observation that this mutant could be the sole reason of secondary resistance.
Most of the initial pre-clinical studies on mutations were conducted in murine cell lines transfected with FLT3 cDNA[32,33,37,45-47]. We and others to have further investigated the molecular mechanisms of acquired resistance to FLT3 inhibitors. Human leukemia cell lines with FLT3 mutations are valuable and relevant models for molecular biology and drug sensitivity studies. MV4-11 and MOLM-14 cell lines were derived from primary AML cells, while MV4-11 has two FLT3-ITD alleles; MOLM-14 harbors one mutant FLT3-ITD allele, while the other allele is wild-type (WT). Leukemia cell line Hb1119 and SEM-K2 were derived from primary ALL (acute lymphoblastic leukemia) cells. Hb1119 harbors FLT3-D836H, whereas SEM-K2 over expresses wild-type FLT3. Piloto et al[49] reported that prolonged coculture of MOLM-14, Hb1119 and SEM-K2 cells with CEP-5214 and CEP-701 respectively leaded to the development of resistant lines including M14(R)5214, M14(R)701, Hb(R)5214, Hb(R)701, SEM(R)5214 and SEM(R)701. They are cross resistant to PKC412 and AG1295, a structurally related FLT3 inhibitor[49]. Although TKIs can inhibit phosphorylation of FLT3 receptor in most of the resistant clones as demonstrated in this study, the downstream Akt and/or MAPK signaling remain activated, thus providing cells sustained survival and proliferative signaling. Acquired N-Ras mutations have been identified in 2 [M14 (R) 5214 and M14 (R) 701] out of the 6 resistant lines. Transducing N-Ras-G12V mutation into MOLM-14 cells results in resistance to CEP-701[49]. So, activation of parallel signaling pathway independent to FLT3 signaling may contribute to secondary resistance in some cases.
Through long-term culture of MV4-11 cell line with the FLT3 inhibitor, ABT-860, a FLT3 inhibitor-resistant line, MV4-11-R was generated[50]. The IC50 of ABT-869 for MV4-11-R line is 52 nmol/L vs 6 nmol/L for the parental MV4-11 cell line. Importantly, other structurally unrelated inhibitors including SU5416, AG1296 and a FLT3 inhibitor III from MERCK, were not effective to MV4-11-R line anymore, suggesting a cross resistant circumstance. Sequencing analysis showed normal sequence of FLT3-TKD in MV4-11-R cells. Western blot and FACS analysis excluded the overexpression of p-FLT3, FLT3 and three multidrug resistance related proteins (MDR, MRP1 and LRP) in this resistant line. But, overexpression of FLT3LG and Survivin was demonstrated at the both transcript and protein level. Down-regulation of suppressor of cytokine signaling (SOCS) proteins (negative regulators of STAT pathways) was also observed in the presence of overactivation of the STAT1, STAT3 and STAT5 pathways in this resistant line. In conclusion, our findings show that overactivation of STAT pathways and subsequently increased expressions of surviving genes are the main mechanism of resistance to FLT3 inhibitors. A total of 9 main studies regarding to primary and acquired resistance is summarized in Table 1.
| Ref. | Disease model (method and material) | Mechanisms of resistance | |
| Primary resistance | Grundler et al[32], 2003 | Site-directed mutagenesis, murine Ba/F3 | TKD mutation, deletion or insertion |
| Clark et al[33], 2004 | Site-directed mutagenesis, murine Ba/F3 | TKD mutation | |
| Bagrintseva et al[37], 2005 | Site-directed mutagenesis, murine Ba/F3 | ITD-TKD mutation, Bcl-xL overexpression | |
| Acquired resistance | Weisberg et al[45], 2002 | Coculture with PKC412, murine Ba/F3-FLT3-ITD | FLT3 protein overexpression |
| Bagrintseva et al[46], 2004 | Coculture with SU5614, murine Ba/F3-FLT3-ITD | ITD-TKD mutation, FLT3 protein overexpression | |
| Cools et al[47], 2004 | Random PCR mutagenesis, murine Ba/F3-FLT3-ITD | ITD-TKD mutation | |
| Heidel et al[48], 2006 | PKC412 clinical trial, relapsed AML with ITD | ITD-TKD (N676K) mutation | |
| Piloto et al[49], 2007 | Coculture with CEP-5214 and CEP-701, human MOLM-14, Hb1119 and SEM-K2 | RTK amplification, N-Ras mutation | |
| Zhou et al[50], 2009 | Coculture with ABT-869, human MV4-11 | Overactivation of STAT, overexpression of Survivin |
The understanding of molecular mechanisms of primary and secondary resistance to FLT3 inhibitors (Figure 1) provides the foundation for establishing strategies to conquer or reduce resistance. Combination of FLT3 inhibitors with cytotoxic drugs or other small molecule inhibitors targeting different pathways has been extensively searched and tested in vitro, in murine xenograft models and some in clinical trials.
We and other investigators have demonstrated that combination of FLT3 inhibitors with conventional chemotherapy drugs, such as cytarabine and doxorubicin, can achieve synergistic effect[30,51-54]. The optimal combination sequence should start with chemotherapy, followed by FLT3 inhibitors to maximize synergism and potential to reduce and/or overcome resistance[30,53].
Combination of FLT3 inhibitors with a spectrum of small molecules inhibitors targeting downstream or independent signaling pathways have been evaluated in pre-clinical studies and showed early promises. Rapamycin, an mTOR inhibitor, sensitizes not only Imatinib-resistant BCR-ABL positive cells[55] but also TKI-resistant Ba/FLT3 dual mutant (ITD and TKD) cell[46,55]. Approaches targeting cellular apoptosis machinery also have been explored. BH3 mimetic ABT-737, a potent inhibitor of anti-apoptotic Bcl2, effectively neutralizes resistance to FLT3 inhibitors in primary AML blasts[56]. The proapoptotic inhibitor LBW242, a Smac (one member of the inhibitor of apoptosis, IAP) mimetic, can overcome resistance to PKC412 when used in combination with PKC412[57]. Combination of FLT3 inhibitor GTP14564 with a HSP90 inhibitor, 17-allylamino-17-demethoxygeldanamycin (17-AAG), produces synergism via STAT5 pathway[58]. Concurrent treatment with histone deacetylase inhibitor (HDACi) LAQ824 and PCK412 can synergistically induced apoptosis in human cell line and primary AML samples with FLT3 mutations[59]. We demonstrate that either treatment with IDR E804, an inhibitor of CDKs and the SRC-STAT pathway, or targeting Survivin by shRNA or a dominant-negative vector (survivin-T34A) sensitize MV4-11-R to ABT-869 induce apoptosis[50].
Other compounds such as bis(1H-indol-2-yl) methanone Cpd.98, Cpd.102 and Sorafenib (B-Raf inhibitor) also overcome resistance to FLT3 inhibitors[60,61]. Downregulation of FLT3 expression by RNAi increases sensitivity to FLT3 inhibitor MLN518 in human AML cell lines, a potential approach to override resistance[62]. The PIM family of serine/threonine kinases (PIM-1, -2 and -3) has been shown to be cytoprotective[63]. Constitutively activated FLT3 signaling up-regulates the PIM-1 expression via STAT5 pathway, which results in phosphorylation of BAD protein (pSer112 and pSer136), exerting anti-apoptotic effect[63,64]. PIM-2 also phosphorylates BAD at Ser-112, blocking BAD-inducing cell death[65]. Silencing PIM-2 or PIM-1 sensitizes resistant cells to FLT3 inhibitors[66]. IMC-EB10, an anti-FLT3 monoclonal antibody, is still effective in FLT3-TKI resistant clones, because it mediates antibody-dependent, cell-mediated cytotoxicity (ADCC) which is independent of the FLT3-ITD signaling pathway[49].
Primary and secondary resistance to TKI therapy is challenging issue in modern anti-cancer warfare for various cancers including AML. At present time, monotherapy using FLT3 inhibitors showed limited benefit in relapsed AML clinical trials. We now began to better understand the molecular mechanisms of resistance in FLT3 targeting. Ongoing early phase clinical trials are important to further shed light on various potential mechanisms of resistance, and will eventually facilitate better strategies to prevent and overcome resistance. Sequel combination of FLT3 inhibitors with chemotherapy or other small molecule inhibitors targeting mTOR, HDAC, HSP90, STAT3, Bcl2, PIM family, IAPs (Survivin and Smac) and others are ongoing strategies. The correlative studies with these ongoing trials for identifying resistance mechanisms among trial patients, will help investigators in refining the design for next generation trial protocols. In addition, by determining “oncogenic signature” of each patient prior to treatment should guide the proper choice of most efficient combinations targeting the specific “oncogenic signature” individually.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: Singapore
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Chiang TA, Gadbail AR, Ghorbian S, Morelli F S- Editor: Ji FF L- Editor: A E- Editor: Wu YXJ
| 1. | de Necochea-Campion R, Shouse GP, Zhou Q, Mirshahidi S, Chen CS. Aberrant splicing and drug resistance in AML. J Hematol Oncol. 2016;9:85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 2. | Hassan C, Afshinnekoo E, Li S, Wu S, Mason CE. Genetic and epigenetic heterogeneity and the impact on cancer relapse. Exp Hematol. 2017;54:26-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 3. | Zhou J, Chooi JY, Ching YQ, Quah JY, Toh SH, Ng Y, Tan TZ, Chng WJ. NF-κB promotes the stem-like properties of leukemia cells by activation of LIN28B. World J Stem Cells. 2018;10:34-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Zhu X, Ma Y, Liu D. Novel agents and regimens for acute myeloid leukemia: 2009 ASH annual meeting highlights. J Hematol Oncol. 2010;3:17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 5. | Shah K, Curtin BF, Chu C, Hwang D, Flasar MH, von Rosenvinge E. Characteristics of Clostridium difficile infection in patients hospitalized with myelodysplastic syndrome or acute myelogenous leukemia. World J Clin Oncol. 2017;8:398-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Medinger M, Passweg JR. Acute myeloid leukaemia genomics. Br J Haematol. 2017;179:530-542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 7. | Bullinger L, Döhner K, Döhner H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J Clin Oncol. 2017;35:934-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 303] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 8. | Zhou J, Lu X, Tan TZ, Chng WJ. X-linked inhibitor of apoptosis inhibition sensitizes acute myeloid leukemia cell response to TRAIL and chemotherapy through potentiated induction of proapoptotic machinery. Mol Oncol. 2018;12:33-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Takahashi S. Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: biology and therapeutic implications. J Hematol Oncol. 2011;4:13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 10. | Gregory TK, Wald D, Chen Y, Vermaat JM, Xiong Y, Tse W. Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J Hematol Oncol. 2009;2:23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 11. | Zhou J, Chan ZL, Bi C, Lu X, Chong PS, Chooi JY, Cheong LL, Liu SC, Ching YQ, Zhou Y. LIN28B Activation by PRL-3 Promotes Leukemogenesis and a Stem Cell-like Transcriptional Program in AML. Mol Cancer Res. 2017;15:294-303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, Sonoda Y, Fujimoto T, Misawa S. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911-1918. [PubMed] [Cited in This Article: ] |
| 13. | Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, Walker H, Wheatley K, Bowen DT, Burnett AK. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752-1759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1139] [Cited by in F6Publishing: 1135] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 14. | Leung AY, Man CH, Kwong YL. FLT3 inhibition: a moving and evolving target in acute myeloid leukaemia. Leukemia. 2013;27:260-268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 15. | Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, Wermke M, Bornhäuser M, Ritter M, Neubauer A. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326-4335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1284] [Cited by in F6Publishing: 1291] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 16. | Minami Y, Yamamoto K, Kiyoi H, Ueda R, Saito H, Naoe T. Different antiapoptotic pathways between wild-type and mutated FLT3: insights into therapeutic targets in leukemia. Blood. 2003;102:2969-2975. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin Cancer Res. 2003;9:2140-2150. [PubMed] [Cited in This Article: ] |
| 18. | Zhou J, Goh BC, Albert DH, Chen CS. ABT-869, a promising multi-targeted tyrosine kinase inhibitor: from bench to bedside. J Hematol Oncol. 2009;2:33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Broekman F, Giovannetti E, Peters GJ. Tyrosine kinase inhibitors: Multi-targeted or single-targeted? World J Clin Oncol. 2011;2:80-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 148] [Cited by in F6Publishing: 146] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 20. | Choudhary C, Schwäble J, Brandts C, Tickenbrock L, Sargin B, Kindler T, Fischer T, Berdel WE, Müller-Tidow C, Serve H. AML-associated Flt3 kinase domain mutations show signal transduction differences compared with Flt3 ITD mutations. Blood. 2005;106:265-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 194] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Zheng R, Friedman AD, Levis M, Li L, Weir EG, Small D. Internal tandem duplication mutation of FLT3 blocks myeloid differentiation through suppression of C/EBPalpha expression. Blood. 2004;103:1883-1890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 121] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Chen Y, Pan Y, Guo Y, Zhao W, Ho WT, Wang J, Xu M, Yang FC, Zhao ZJ. Tyrosine kinase inhibitors targeting FLT3 in the treatment of acute myeloid leukemia. Stem Cell Investig. 2017;4:48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 23. | Park JE, Yuen HF, Zhou JB, Al-Aidaroos AQ, Guo K, Valk PJ, Zhang SD, Chng WJ, Hong CW, Mills K. Oncogenic roles of PRL-3 in FLT3-ITD induced acute myeloid leukaemia. EMBO Mol Med. 2013;5:1351-1366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 24. | Sternberg DW, Licht JD. Therapeutic intervention in leukemias that express the activated fms-like tyrosine kinase 3 (FLT3): opportunities and challenges. Curr Opin Hematol. 2005;12:7-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 25. | Zhou J, Bi C, Chng WJ, Cheong LL, Liu SC, Mahara S, Tay KG, Zeng Q, Li J, Guo K. PRL-3, a metastasis associated tyrosine phosphatase, is involved in FLT3-ITD signaling and implicated in anti-AML therapy. PLoS One. 2011;6:e19798. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 26. | Wei AH, Tiong IS. Midostaurin, enasidenib, CPX-351, gemtuzumab ozogamicin, and venetoclax bring new hope to AML. Blood. 2017;130:2469-2474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 27. | Larrosa-Garcia M, Baer MR. FLT3 Inhibitors in Acute Myeloid Leukemia: Current Status and Future Directions. Mol Cancer Ther. 2017;16:991-1001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 196] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 28. | Lancet JE. New agents: great expectations not realized. Best Pract Res Clin Haematol. 2013;26:269-274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Swords R, Freeman C, Giles F. Targeting the FMS-like tyrosine kinase 3 in acute myeloid leukemia. Leukemia. 2012;26:2176-2185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 107] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 30. | Zhou J, Pan M, Xie Z, Loh SL, Bi C, Tai YC, Lilly M, Lim YP, Han JH, Glaser KB. Synergistic antileukemic effects between ABT-869 and chemotherapy involve downregulation of cell cycle-regulated genes and c-Mos-mediated MAPK pathway. Leukemia. 2008;22:138-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Melo JV, Chuah C. Novel agents in CML therapy: tyrosine kinase inhibitors and beyond. Hematology Am Soc Hematol Educ Program. 2008;427-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 32. | Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102:646-651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 33. | Clark JJ, Cools J, Curley DP, Yu JC, Lokker NA, Giese NA, Gilliland DG. Variable sensitivity of FLT3 activation loop mutations to the small molecule tyrosine kinase inhibitor MLN518. Blood. 2004;104:2867-2872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 63] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Chen W, Jones D, Medeiros LJ, Luthra R, Lin P. Acute myeloid leukaemia with FLT3 gene mutations of both internal tandem duplication and point mutation type. Br J Haematol. 2005;130:726-728. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 35. | Georgiou G, Karali V, Zouvelou C, Kyriakou E, Dimou M, Chrisochoou S, Greka P, Dufexis D, Vervesou E, Dimitriadou E. Serial determination of FLT3 mutations in myelodysplastic syndrome patients at diagnosis, follow up or acute myeloid leukaemia transformation: incidence and their prognostic significance. Br J Haematol. 2006;134:302-306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 36. | Moreno I, Martín G, Bolufer P, Barragán E, Rueda E, Román J, Fernández P, León P, Mena A, Cervera J. Incidence and prognostic value of FLT3 internal tandem duplication and D835 mutations in acute myeloid leukemia. Haematologica. 2003;88:19-24. [PubMed] [Cited in This Article: ] |
| 37. | Bagrintseva K, Geisenhof S, Kern R, Eichenlaub S, Reindl C, Ellwart JW, Hiddemann W, Spiekermann K. FLT3-ITD-TKD dual mutants associated with AML confer resistance to FLT3 PTK inhibitors and cytotoxic agents by overexpression of Bcl-x(L). Blood. 2005;105:3679-3685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 80] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 38. | Irish JM, Anensen N, Hovland R, Skavland J, Børresen-Dale AL, Bruserud O, Nolan GP, Gjertsen BT. Flt3 Y591 duplication and Bcl-2 overexpression are detected in acute myeloid leukemia cells with high levels of phosphorylated wild-type p53. Blood. 2007;109:2589-2596. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Hunter HM, Pallis M, Seedhouse CH, Grundy M, Gray C, Russell NH. The expression of P-glycoprotein in AML cells with FLT3 internal tandem duplications is associated with reduced apoptosis in response to FLT3 inhibitors. Br J Haematol. 2004;127:26-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 40. | Galimberti S, Rossi A, Palumbo GA, Morabito F, Guerrini F, Vincelli I, Fazzi R, Santini V, Petrini M. FLT3 mutations do not influence MDR-1 gene expression in acute myeloid leukemia. Anticancer Res. 2003;23:3419-3426. [PubMed] [Cited in This Article: ] |
| 41. | Siendones E, Barbarroja N, Torres LA, Buendía P, Velasco F, Dorado G, Torres A, López-Pedrera C. Inhibition of Flt3-activating mutations does not prevent constitutive activation of ERK/Akt/STAT pathways in some AML cells: a possible cause for the limited effectiveness of monotherapy with small-molecule inhibitors. Hematol Oncol. 2007;25:30-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 42. | Mony U, Jawad M, Seedhouse C, Russell N, Pallis M. Resistance to FLT3 inhibition in an in vitro model of primary AML cells with a stem cell phenotype in a defined microenvironment. Leukemia. 2008;22:1395-1401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 43. | Pollard JA, Alonzo TA, Gerbing RB, Woods WG, Lange BJ, Sweetser DA, Radich JP, Bernstein ID, Meshinchi S. FLT3 internal tandem duplication in CD34+/CD33- precursors predicts poor outcome in acute myeloid leukemia. Blood. 2006;108:2764-2769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 44. | Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876-880. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2357] [Cited by in F6Publishing: 2280] [Article Influence: 99.1] [Reference Citation Analysis (0)] |
| 45. | Weisberg E, Boulton C, Kelly LM, Manley P, Fabbro D, Meyer T, Gilliland DG, Griffin JD. Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412. Cancer Cell. 2002;1:433-443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 463] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 46. | Bagrintseva K, Schwab R, Kohl TM, Schnittger S, Eichenlaub S, Ellwart JW, Hiddemann W, Spiekermann K. Mutations in the tyrosine kinase domain of FLT3 define a new molecular mechanism of acquired drug resistance to PTK inhibitors in FLT3-ITD-transformed hematopoietic cells. Blood. 2004;103:2266-2275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 47. | Cools J, Mentens N, Furet P, Fabbro D, Clark JJ, Griffin JD, Marynen P, Gilliland DG. Prediction of resistance to small molecule FLT3 inhibitors: implications for molecularly targeted therapy of acute leukemia. Cancer Res. 2004;64:6385-6389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 144] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 48. | Heidel F, Solem FK, Breitenbuecher F, Lipka DB, Kasper S, Thiede MH, Brandts C, Serve H, Roesel J, Giles F. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293-300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 222] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 49. | Piloto O, Wright M, Brown P, Kim KT, Levis M, Small D. Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood. 2007;109:1643-1652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 136] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 50. | Zhou J, Bi C, Janakakumara JV, Liu SC, Chng WJ, Tay KG, Poon LF, Xie Z, Palaniyandi S, Yu H. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood. 2009;113:4052-4062. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 120] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 51. | Aleskog A, Höglund M, Pettersson J, Hermansson M, Larsson R, Lindhagen E. In vitro activity of the flt3-inhibitor su5614 and standard cytotoxic agents in tumour cells from patients with wild type and mutated flt3 acute myeloid leukaemia. Leuk Res. 2005;29:1079-1081. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 52. | Furukawa Y, Vu HA, Akutsu M, Odgerel T, Izumi T, Tsunoda S, Matsuo Y, Kirito K, Sato Y, Mano H. Divergent cytotoxic effects of PKC412 in combination with conventional antileukemic agents in FLT3 mutation-positive versus -negative leukemia cell lines. Leukemia. 2007;21:1005-1014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 53. | Levis M, Pham R, Smith BD, Small D. In vitro studies of a FLT3 inhibitor combined with chemotherapy: sequence of administration is important to achieve synergistic cytotoxic effects. Blood. 2004;104:1145-1150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 182] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 54. | Yee KW, Schittenhelm M, O’Farrell AM, Town AR, McGreevey L, Bainbridge T, Cherrington JM, Heinrich MC. Synergistic effect of SU11248 with cytarabine or daunorubicin on FLT3 ITD-positive leukemic cells. Blood. 2004;104:4202-4209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 101] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 55. | Mohi MG, Boulton C, Gu TL, Sternberg DW, Neuberg D, Griffin JD, Gilliland DG, Neel BG. Combination of rapamycin and protein tyrosine kinase (PTK) inhibitors for the treatment of leukemias caused by oncogenic PTKs. Proc Natl Acad Sci USA. 2004;101:3130-3135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 200] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 56. | Kohl TM, Hellinger C, Ahmed F, Buske C, Hiddemann W, Bohlander SK, Spiekermann K. BH3 mimetic ABT-737 neutralizes resistance to FLT3 inhibitor treatment mediated by FLT3-independent expression of BCL2 in primary AML blasts. Leukemia. 2007;21:1763-1772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 86] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 57. | Weisberg E, Kung AL, Wright RD, Moreno D, Catley L, Ray A, Zawel L, Tran M, Cools J, Gilliland G. Potentiation of antileukemic therapies by Smac mimetic, LBW242: effects on mutant FLT3-expressing cells. Mol Cancer Ther. 2007;6:1951-1961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 58. | Yao Q, Nishiuchi R, Kitamura T, Kersey JH. Human leukemias with mutated FLT3 kinase are synergistically sensitive to FLT3 and Hsp90 inhibitors: the key role of the STAT5 signal transduction pathway. Leukemia. 2005;19:1605-1612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 59. | Bali P, George P, Cohen P, Tao J, Guo F, Sigua C, Vishvanath A, Scuto A, Annavarapu S, Fiskus W. Superior activity of the combination of histone deacetylase inhibitor LAQ824 and the FLT-3 kinase inhibitor PKC412 against human acute myelogenous leukemia cells with mutant FLT-3. Clin Cancer Res. 2004;10:4991-4997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 60. | Heidel F, Lipka DB, Mirea FK, Mahboobi S, Grundler R, Kancha RK, Duyster J, Naumann M, Huber C, Böhmer FD. Bis(1H-indol-2-yl)methanones are effective inhibitors of FLT3-ITD tyrosine kinase and partially overcome resistance to PKC412A in vitro. Br J Haematol. 2009;144:865-874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 61. | Lierman E, Lahortiga I, Van Miegroet H, Mentens N, Marynen P, Cools J. The ability of sorafenib to inhibit oncogenic PDGFRbeta and FLT3 mutants and overcome resistance to other small molecule inhibitors. Haematologica. 2007;92:27-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 66] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 62. | Walters DK, Stoffregen EP, Heinrich MC, Deininger MW, Druker BJ. RNAi-induced down-regulation of FLT3 expression in AML cell lines increases sensitivity to MLN518. Blood. 2005;105:2952-2954. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 30] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 63. | Kim KT, Baird K, Ahn JY, Meltzer P, Lilly M, Levis M, Small D. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood. 2005;105:1759-1767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 186] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 64. | Kim KT, Levis M, Small D. Constitutively activated FLT3 phosphorylates BAD partially through pim-1. Br J Haematol. 2006;134:500-509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 72] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 65. | Yan B, Zemskova M, Holder S, Chin V, Kraft A, Koskinen PJ, Lilly M. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem. 2003;278:45358-45367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 211] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 66. | Adam M, Pogacic V, Bendit M, Chappuis R, Nawijn MC, Duyster J, Fox CJ, Thompson CB, Cools J, Schwaller J. Targeting PIM kinases impairs survival of hematopoietic cells transformed by kinase inhibitor-sensitive and kinase inhibitor-resistant forms of Fms-like tyrosine kinase 3 and BCR/ABL. Cancer Res. 2006;66:3828-3835. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 88] [Article Influence: 4.9] [Reference Citation Analysis (0)] |