Bruchid Resistance Study through Bulked Segregant Analysis: Used as a Preliminary Step for Next-Generation Sequencing
DOI:
https://doi.org/10.48048/tis.2022.3975Keywords:
Bruchid resistance, RILs, Bulked segregant analysis, Molecular markersAbstract
Seed beetles, commonly known as bruchids, are serious insect pests of Indian pulses inflicting high damage during storage. Earlier, a bruchid resistant rice bean landrace (TNAU Red) from Manipur and a susceptible mung bean variety (VRMGg1) from Tamil Nadu were reported and their inter-specific cross was made in Tamil Nadu Agricultural University. In the present study, seeds from the segregating populations (recombinant inbred lines, RILs) of VRM (Gg)1 and TNAU Red were evaluated for bruchid Callosobruchus maculatus Fabricius resistance following standard methods under laboratory conditions. Bulked segregant analysis (BSA) is an efficient method for quick identification of molecular markers linked to any specific gene or genomic region. BSA significantly reduces the scale and cost by simplifying the procedure compared to conventional method of analyzing the entire population. BSA was carried out using 3 marker systems with an attempt to identify markers associated with the present trait of interest. Two DNA bulks namely resistant bulk (RB) and susceptible bulk (SB) were grouped by pooling equal amount of DNA from 5 each of resistant and susceptible RILs based on bruchid responses such as seed damage (%), mean developmental days (MDP) and adult emergence from seeds. Marker analysis carried out with 41 polymorphic SSRs showed skewed segregation distortion towards the susceptible parent. However, out of 9 RAPDs and 10 ISSRs, 2 RAPD (OPB08 and OPX04) and 1 ISSR (UBC810) primers amplified an allele at 200 - 300 and 300 - 400 bp, respectively, in resistant parent and resistant bulk but not in susceptible parent and susceptible bulk. The amplification of resistant parent allele in resistant bulk may indicate the presence of bruchid resistant gene(s) in the resistant RILs. This experiment was aimed at verifying the presence of bruchid resistant lines carrying resistant gene(s) prior to further association mapping or QTL mapping studies. Further, such type of study can be applied as a preliminary step in NGS technology.
HIGHLIGHTS
- Bruchids are the most damaging storage insect in pulses and other legumes causing great economic losses
- Bulk segregant analysis (BSA) conducted using three marker systems in an attempt to identify markers associated with bruchid resistance
- Categorization of RILs based on bruchid responses such as seed damage (%), mean developmental days (MDP), adult emergence from seeds and scoring done
- Marker analysis using BSA useful for verification of the presence of bruchid resistant lines carrying resistant gene(s) prior to further advanced studies such as QTL mapping, NGS, etc.
GRAPHICAL ABSTRACT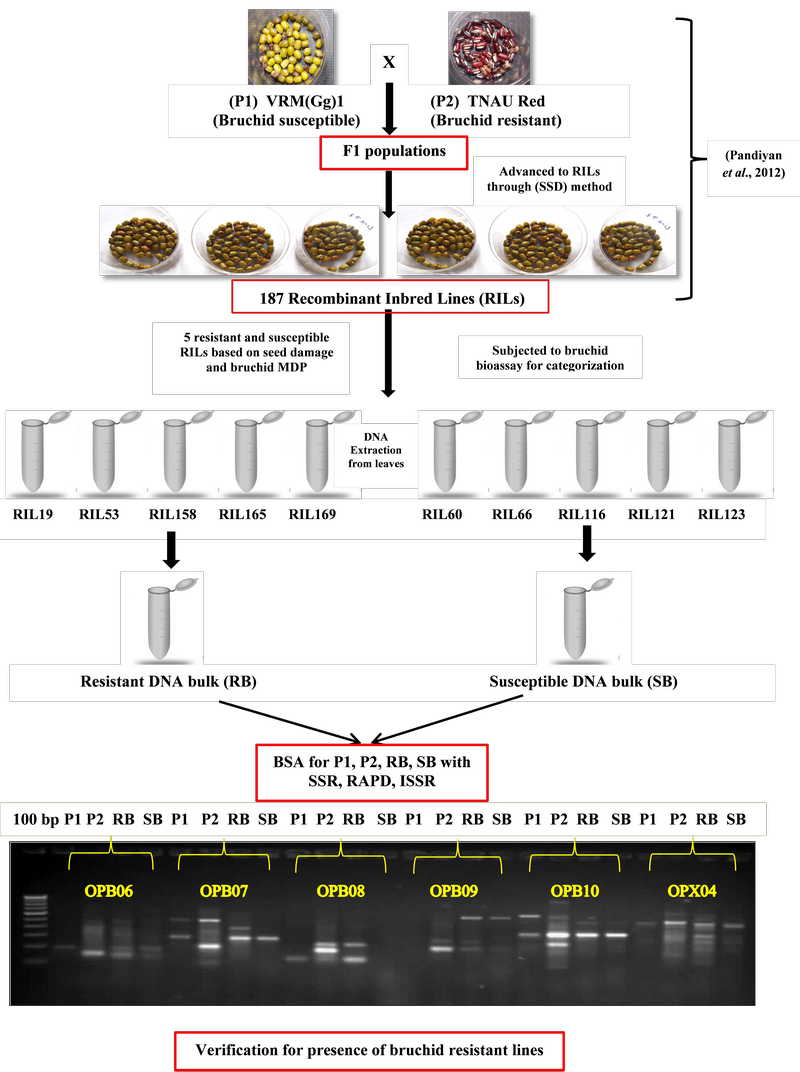
Downloads
Metrics
References
N Tomooka, DA Vaughan, H Moss and N Maxted. The Asian Vigna: Genus Vigna subgenus Ceratotropis genetic resources. Dordrecht, Netherlands, 2002, p. 270.
RK Arora, KPS Chandel, BS Joshi and KC Pant. Rice Bean: Tribal pulse of eastern India. Econ. Bot. 1980; 34, 260-3.
D Seram, N Senthil, M Pandiyan and JS Kennedy. Resistance determination of a South Indian bruchid strain against rice bean landraces of Manipur (India). J. Stored Prod. Res. 2016; 69, 199-206.
R Radha and P Susheela. Studies on the life history and ovipositional Preference of Callosobruchus maculatus reared on different pulses. Res. J. Anim. Vet. Fishery Sci. 2014; 2, 1-5.
D Seram, N Senthil and JS Kennedy. Comparative susceptibility and life per for mance studies of Callosobruchus Maculatus F. on Vigna Radiata (L.) Wilcezk, seeds. Prog. Res. Int. J. 2015; 10, 662-5.
D Seram, S Mohan, JS Kennedy and N Senthil. Development and damage assessment of the storage beetle, Callosobruchus maculatus (Thanjavur and Coimbatore strain) under normal and controlled conditions. In: Proceedings of the 10th International Conference on Controlled Atmosphere and Fumigation in Stored Products, Winnipeg, Canada. 2016.
JH Appleby and PF Credland. Variation in responses to susceptible and resistant cowpeas among West African populations of Callosobruchus maculatus (Coleoptera: Bruchidae). J. Econ. Entomol. 2003; 96, 489-502.
S Vir. Ovipositional response and development of Callosobruchus maculatus (Fab.) on different varieties of cowpea. Bull. Grain Tech. 1980; 18, 200-3.
GB Swella and DMK Mushobozy. Comparative susceptibility of different legume seeds to infestation by cowpea bruchid Callosobruchus maculatus (F.) (Coleoptera: Chrysomelidae). Plant Protect. Sci. 2009; 45, 19-24.
N Tomooka, K Kashiwaba, DA Vaughan, M Ishimoto and Y Egawa. The effectiveness of evaluating wild species: Searching for sources of resistance to bruchid beetles in the genus Vigna subgenus Ceratotropis. Euphytica 2000; 115, 27-41.
CJ Lambrides and BC Imrie. Susceptibility of mungbean varieties to the bruchid species Callosobruchus maculatus (F.), C-phaseoli (Gyll.), C-chinensis (L.), and Acanthoscelides obtectus (Say.) (Coleoptera: Chrysomelidae). Aust. J. Agr. Res. 2000; 51, 85-9.
MLR Macedo, LBDS Andrade, RA Moraes and J Xavier. Vicilin variants and the resistance of cowpea (Vigna unguiculata) seeds to the cowpea weevil (Callosobruchus maculatus). Comp. Biochem. Physiol. C Comp. Pharmacol. 1993; 105, 89-4.
K Kashiwaba and N Tomooka. Isolation and identification of factors responsible for resistance to bruchid in rice bean. Annual Report, 2002, p. 8-9.
M Sudha, P Anusuya, NG Mahadev, A Karthikeyan, P Nagarajan, M Raveendran and P Balasubramanian. Molecular studies on mungbean (Vigna radiata (L.) Wilczek) and ricebean (Vigna umbellata (Thunb.)) interspecific hybridisation for Mungbean yellow mosaic virus resistance and development of species-specific SCAR marker for ricebean. Arch. Phytopathology Plant Protect. 2013; 46, 503-17.
N Tomooka, C Lairungreang, P Nakeeraks, Y Egawa and C Thavarasook. Development of bruchid-resistant mungbean line using wild mungbean germplasm in Thailand. Plant Breed. 1992; 109, 60-6.
P Somta, C Ammaranan, A Peter, PAC Ooi and P Srinives. Inheritance of seed resistance to bruchids in cultivated mung bean (Vigna radiata). Euphytica 2007; 1-2, 47-55.
R Schafleitner, S Huang, S Chu, J Yen, C Lin, M Yan, B Krishnan, M Liu, H Lo, C Chen, LO Chen, D Wu, TT Bui, S Ramasamy, C Tung and R Nair. Identification of single nucleotide polymorphism markers associated with resistance to bruchids (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) and cultivated V. radiata through genotyping by sequencing and quantitative trait locus analysis. BMC Plant Biol. 2016; 16, 159.
ND Young, L Kumar, DM Hautea, D Danesh, NS Talekar, S Shanmugasundarum and DH Kim. RFLP mapping of a major bruchid resistance gene in mungbean (Vigna radiata, L. Wilczek). Theor. Appl. Genet. 1992; 84, 839-44.
A Kaga and M Ishimoto. Genetic localization of a bruchid resistance gene and its relationship to insecticidal cyclopeptide alkaloids, the vignatic acids, in mungbean (Vigna radiata L. Wilczek). Mol. Genet. Genom. 1998; 258, 378-84.
CJ Lambrides, RJ Lawn, ID Godwin, J Manners and BC Imrie. Two genetic linkage maps of mungbean using RFLP and RAPD markers. Aust. J. Agr. Res. 2000; 51, 415-25.
T Isemura, A Kaga, S Tabata, P Somta, P Srinives, T Shimizu, U Jo, DA Vaughan and N Tomooka. Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLos ONE 2012; 7, e41304.
S Chotechung, P Somta, J Chen, T Yimram, X Chen and P Srinives. A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 2016; 129, 1673-83.
P Somta, A Kaga, N Tomooka, K Kashiwaba, T Isemura, B Chaitieng, P Srinives and DA Vaughan. Development of an interspecific Vigna linkage map between Vigna umbellata (Thunb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi & Ohashi and its use in analysis of bruchid resistance and comparative genomics. Plant. Breed. 2006; 125, 77-84.
RW Michelmore, I Paran and RV Kesseli. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA. 1991; 88, 9828-32.
Y Han, P Lv, S Hou, S Li, G Ji, X Ma, R Du and G Liu. Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in Sorghum (Sorghum bicolor L. Moench). PLos ONE 2015; 10, e0127065.
M Pandiyan, N Senthil, N Ramamoorthi, AR Muthiah, N Tomooka, V Duncan and T Jayaraj. Interspecific hybridization of Vigna radiata x 13 wild Vigna species for developing MYMV donar. Electron. J. Plant Breed. 2012; 1, 600-10.
MK Mathivathana, SM Samyuktha, RD Priya, I Mariyammal, P Bharathi, N Jagadeeshselvam, A Karthikeyan, M Sudha, M Pandiyan, G Karthikeyan, C Vanniarajan, M Raveendran and N Senthil. Association and path analysis of yield and yield components in the RIL population of Vigna radiata × Vigna umbellata. Int. J. Curr. Microbiol. App. Sci. 2018; 7, 1231-5.
MK Mathivathana, J Murukarthick, A Karthikeyan, W Jang, M Dhasarathan, N Jagadeeshselvam, M Sudha, C Vanniarajan, G Karthikeyan, TJ Yang, M Raveendran, M Pandiyan and N Senthil. Detection of QTLs associated with mungbean yellow mosaic virus (MYMV) resistance using the interspecific cross of Vigna radiata × Vigna umbellata. J. Appl. Genet. 2019; 60, 255-68.
L Sun, X Cheng, S Wang, L Wang, C Liu, L Mei and N Xu. Heredity analysis and gene mapping of bruchid resistance of a mung bean cultivar C2709. Agr. Sci. China 2008; 7, 672-7.
S Manickam. DNA isolation protocol for Vigna radiata with free of phenolics. Nat. Protocol. 2009, https://doi.org/10.1038/nprot.2009.167.
M Pandiyan, N Ramamoorthi, SK Ganesh, S Jebaraj, P Pagarajan and P Balasubramanian. Broadening the genetic base and introgression of MYMV resistance and yield improvement through unexplored genes from wild relatives in mungbean. Plant Mutat. Res. 2008; 2, 33-8.
B Burr and FA Burr. Zein synthesis in maize endosperm by polyribosomes attached to protein bodies. Proc. Nat. Acad. Sci. 1976; 73, 515-19.
AH Paterson, ES Lander, JD Hewitt, S Peterson, SE Lincoln and SD Tanksley. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 1998; 335, 721-6.
N Jones, H Ougham and H Thomas. Markers and mapping: We are all geneticists now. New Phytol. 1997; 137, 165-77.
L Mandal, SK Verma, S Sasmal and J Katara. Potential applications of molecular markers in plant. Curr. Trends Biomed. Eng. Biosci. 2018; 12, 555844.
A Kaga, DA Vaughan and N Tomooka. Molecular markers in Vigna improvement: Understanding and using gene pools. In: H Lörz and G Wenzel (Eds.). molecular marker systems in plant breeding and crop improvement. Biotechnology in Agriculture and Forestry, Heidelberg, Germany, 2004, p. 171-87.
HM Chen, CA Liu, CG Kuo, CM Chien, HC Sun, CC Huang, YC Lin and HM Ku. Development of a molecular marker for a bruchid (Callosobruchus chinensis L.) resistance gene in mungbean. Euphytica 2007; 157, 113-22.
MG Hong, KH Kim, J Ku, J Jeong, MJ Seo, CH Park, YH Kim, HS Kim, YK Kim, S Baek, DY Kim, SK Park, SL Kim and J Moon. Inheritance and quantitative trait loci analysis of resistance genes to bruchid and bean bug in mungbean (Vigna radiata L. Wilczek). Plant Breed. Biotech. 2015; 3, 39-46.
MP Reddy, N Sarla and EA Siddiq. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002; 128, 9-17.
ID Godwin, EAB Aitken and LW Smith. Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis 2005; 18, 1524-8.
I Mariyammal, D Seram, SM Samyuktha, A Karthikeyan, M Dhasarathan, J Murukarthick, JS Kennedy, D Malarvizhi, TJ Yang, M Pandiyan and N Senthil. QTL mapping in Vigna radiata × Vigna umbellata population uncovers major genomic regions associated with bruchid resistance. Mol. Breed. 2019; 39, 110.
J Song, Z Li, Z Liu, Y Guo and LJ Qiu. Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front. Plant Sci. 2017; 8, 919.
M Trick, NM Adamski, SG Mugford, CC Jiang, M Febrer and C Uauy. Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol. 2012; 12, 14.
H Takagi, A Abe, K Yoshida, S Kosugi, S Natsume, C Mitsuoka, A Uemura, H Utsushi, M Tamiru, S Takuno, H Innan, LM Cano, S Kamoun and R Terauchi. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013; 74, 174-83.
M Livaja, Y Wang, S Wieckhorst, G Haseneyer, M Seidel and V Hahn. BSTA: A targeted approach combines bulked segregant analysis with next-generation sequencing and de novo transcriptome assembly for SNP discovery in sunflower. BMC Genom. 2013; 14, 628.
W Dong, D Wu, G Li, D Wu and Z Wang. Next-generation sequencing from bulked segregant analysis identifies a dwarfism gene in watermelon. Sci. Rep. 2018; 8, 2908.
MPD Silva, AZ Zaccaron, BH Bluhm, JC Rupe, L Wood, LA Mozzoni, RE Mason, S Yingling and A Pereira. Bulked segregant analysis using next-generation sequencing for identification of genetic loci for charcoal rot resistance in soybean. Physiol. Mol. Plant Pathol. 2020; 109, 101440.
Downloads
Published
How to Cite
Issue
Section
License

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.







