Published online Nov 27, 2014. doi: 10.4254/wjh.v6.i11.818
Revised: October 10, 2014
Accepted: October 23, 2014
Published online: November 27, 2014
AIM: To explore the potential usefulness of serum miR-122 and miR-221 as non-invasive diagnostic markers of hepatitis C virus (HCV)-related hepatocellular carcinoma (HCC).
METHODS: This prospective study was conducted on 90 adult patients of both sex with HCV-related chronic liver disease and chronic hepatitis C related HCC. In addition to the 10 healthy control individuals, patients were stratified into; interferon-naïve chronic hepatitis C (CH) (n = 30), post-hepatitis C compensated cirrhosis (LC) (n = 30) and treatment-naïve HCC (n = 30). All patients and controls underwent full clinical assessment and laboratory investigations in addition to the evaluation of the level of serum miRNA expression by RT-PCR.
RESULTS: There was a significant fold change in serum miRNA expression in the different patient groups when compared to normal controls; miR-122 showed significant fold increasing in both CH and HCC and significant fold decrease in LC. On the other hand, miR-221 showed significant fold elevation in both CH and LC groups and significant fold decrease in HCC group (P = 0.01). Comparing fold changes in miRNAs in HCC group vs non HCC group (CH and Cirrhosis), there was non-significant fold elevation in miR-122 (P = 0.21) and significant fold decreasing in miR-221 in HCC vs non-HCC (P = 0.03). ROC curve analysis for miR-221 yielded 87% sensitivity and 40% specificity for the differentiation of HCC patients from non-HCC at a cutoff 1.82.
CONCLUSION: Serum miR-221 has a strong potential to serve as one of the novel non-invasive biomarkers of HCC.
Core tip: In the current study a signature of circulating miRNAs (miR-122 and miR-221) was evaluated. miR-221 was differentially expressed between patients with hepatocellular carcinoma and those without (chronic hepatitis and liver cirrhosis) with lower serum level of miR-221 in former group of patients in comparison to later one. miR-221 yielded 87% sensitivity and 40% specificity in differentiating between both groups at a cutoff 1.82 folds. The present study emphasizes that circulating miR-221 deserves further attention as a potential non-invasive biomarkers for hepatocellular carcinoma.
- Citation: El-Garem H, Ammer A, Shehab H, Shaker O, Anwer M, El-Akel W, Omar H. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol 2014; 6(11): 818-824
- URL: https://www.wjgnet.com/1948-5182/full/v6/i11/818.htm
- DOI: https://dx.doi.org/10.4254/wjh.v6.i11.818
Hepatocellular carcinoma (HCC) is the most common primary liver cancer, and the fifth most common cancer worldwide[1]. The last decade has witnessed a significant rise in the incidence of HCC[2-4] with a specially high incidence reported in Egypt[5]. A direct role of hepatitis C virus (HCV) in hepatocarcinogenesis has been suggested[6]. However, it seems that cirrhosis is the common route through which several risk factors act and induce carcinogenesis[7].
Currently available blood tumor markers are far from optimal. Alfa-fetoprotein (AFP), Lens culinaris agglutinin-reactive AFP (AFP-L3) and des-carboxyprothrombin (DCP) perform poorly in the surveillance mode and early detection of HCC[8]. The Practice Guidelines of the American Association for the Study of Liver Diseases (AASLD) (July 2010) rejected AFP whether for the surveillance or the diagnosis of HCC. This highlights the need for other methods that would be minimally-invasive, simple and reliable for the early detection of HCC.
MicroRNA (miRNA) is a non-coding RNA gene product that negatively controls gene expression by altering the stability or translational efficiency of its target mRNAs[2]. MiRNAs regulate several biological processes, such as cell differentiation, apoptosis and proliferation. miRNAs have been reported to be aberrantly present in cancers whether through up- or down-regulation in neoplastic cells compared with their normal counterparts[3,4]. What makes miRNAs even more interesting is that several recent studies have demonstrated that miRNAs are detectable and stable in plasma and serum[4-6].
The goal of the present study was to evaluate circulating serum miRNAs (miR-122 and miR-221) expression levels in Egyptian patients with HCC as well as in patients with HCV-related chronic liver disease to explore their potential as novel non-invasive markers for diagnosis of HCV-related HCC.
During the period between March and June 2012 serum samples were collected from consecutive HCV-infected patients presenting to our outpatient department: 30 with chronic HCV alone (CH), 30 with HCV-related cirrhosis (LC) and 30 with HCV-related HCC. Serum samples were also collected from 10 age and gender-matched healthy volunteers (defined as those with normal transaminases, normal hepatic ultrasound and negative for HBsAg, HBc-Ab and HCV RNA-PCR). All patients were recruited after a written informed consent and the study protocol was approved by the ethics review committee of Cairo University hospital. Exclusion criteria included: patients with chronic HBV infection or any other identifiable cause for chronic hepatitis other than HCV, previous treatment for HCC or antiviral therapy for HCV and any associated malignancies other than HCC.
For the real-time PCR RNAs were extracted from serum using TRIzol according to the manufacturer’s instruction. The RNA purity was assessed by the RNA concentration and quantified by NanoDrop ND-1000 (Nanodrop, United States). Single-stranded cDNAs were generated using the RT kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s directions (miSCript miRNA PCR system, miRneasy mini kit for miRna extraction, miScript RT II for miRna reverse transcription, miSCript Primer Assay and miSCript SYBR Green PCR Kit for PCR amplification.
PCR quantification experiments were performed with PCR (Applied Biosystems; Foster City, CA) using the SYBR Green PCR Master Mix according to the manufacturer’s protocol. The primers for microRNA-122, -221 and housekeeping gene were supplied by Qiagene, Germany (catalog numbers 3416, 3857 and 33712). The housekeeping miRNA SNORD68 was used as the endogenous control. Fluorescence measurements were made in every cycle and the cycling conditions used were: 95 °C for 30 s, and 40 cycles of 95 °C for 5 s and 60 °C for 34 s.
Expression of miRNAs was reported as ΔCt value. The ΔCt was calculated by subtracting the Ct values of miRNA SNORD68 from the Ct values of the target miRNAs. As there is an inverse correlation between ΔCt and miRNA expression level, lower ΔCt values were associated with increased miRNA. The resulting normalized ΔCt values were used in calculating relative expression values by using 2- Δ(Ct), these values are directly related to the miRNA expression levels. The 2-ΔΔ (Ct) method was used to determine relative-quantitative levels of individual miRNAs.
Patients were categorized into 4 groups; normal, CH, Cirrhosis and HCC. Further comparisons were performed between HCC group and Non-HCC (CH and cirrhosis). Quantitative variables were expressed by mean ± SD or expressed by median and inter quartile range (IQR) for non-parametric data. They were compared by t-student or ANOVA test when appropriate. Qualitative variables were compared by χ2 or Fischer’s exact test when appropriate. AFP levels were transformed into their log values to undergo parametric statistical tests. Receiver operator characteristic (ROC) curves were constructed to assess the value of miRNA in diagnosing HCC and to assess area under the curve (AUROC). AUROC less than 0.60 with P value > 0.05 is considered unreliable for ROC curve. Spearmen and Pearson correlations were done for correlating quantitative variables. In all tests, P value was considered significant if less than 0.05.
This study was conducted on 100 participants stratified into: Group1: thirty patients with HCV-related HCC who were diagnosed according to EASL guidelines 2012; Group2: thirty patients with hepatitis C related liver cirrhosis; Group 3: thirty non-cirrhotic patients with chronic hepatitis C viral infection (CH), while Group 4 included ten age and gender-matched healthy volunteers (defined as those with normal hepatic ultrasound and transaminases and negative for hepatitis B and C by PCR) considered as internal reference.
The demographic and pathologic features of the studied participants are shown in Tables 1 and 2. There was a significant difference between the diseased groups regarding age (P < 0.001). Regarding gender difference; males were predominant in HCV related liver disease patients in the three groups and they represented 83.3%, 70%, 73.3% in HCC, cirrhosis and CH groups respectively with no statistically significant difference between the studied groups (P = 0.60).
| HCC (n = 30) | Cirrhosis (n = 30) | CH (n = 30) | Normal (n = 10) | P value | |
| Age (yr) Mean ± SD | 60.27 ± 8.20C | 55.07 ± 7.35B | 38.20 ± 8.21A | 40.89 ± 16.85A | ≤ 0.001 |
| Gender (male) | 25 (83.3) | 21 (70) | 22 (73.3) | 6 (66.7) | 0.6 |
| Hb (g/dL ) | 11.44 ± 2.85B | 10.53 ± 2.00B | 14.23 ± 1.58A | 14.03 ± 2.48A | < 0.001 |
| WBC × 103/mm3 | 5.73 ± 2.56A | 7.08 ± 3.77A | 6.15 ± 2.13A | 7.38 ± 2.72A | 0.217 |
| Platelets 10/mm3D | 126.00 ± 74.05B | 116.10 ± 68.94B | 228.80 ± 59.74A | 271.3 ± 116.7A | < 0.001 |
| Total bilirubin (0.1-1.2 mg/dL) | 1.88 ± 2.01A | 4.29 ± 6.75B | 0.74 ± 0.26A | 0.79 ± 0.36A | < 0.001 |
| ALT (0-42 IU/L) | 66.59 ± 44.59B | 32.15 ± 23.59A | 66.78 ± 36.56B | 29.16 ± 19.96A | < 0.001 |
| AST (0-42 IU/L) | 119.99 ± 56.12B | 63.81 ± 39.49A | 59.67 ± 45.04A | 45.64 ± 54.32A | < 0.001 |
| ALP (0-290 IU/L)D | 394.8 ± 282.28A | 304.89 ± 191.85A | 203.58 ± 82.37AB | 198.8 ± 139.1B | 0.003 |
| Albumin (3.5-5.5 g/dL)D | 3.16 ± 0.40C | 2.49 ± 0.54B | 4.22 ± 0.36A | 4.09 ± 0.92A | < 0.001 |
| PC % | 69.63 ± 16.25C | 51.58 ± 17.12B | 88.24 ± 10.89A | 97.63 ± 11.77A | < 0.001 |
| AFP log10 ng/dL | 2.50 ± 1.19B | 0.79 ± 0.54A | 0.59 ± 0.38A | NA | < 0.001 |
| Parameter | Number (%) | |
| AFP level (0-10) | Normal | 4 (13.4%) |
| Elevated | 26 (86.6%) | |
| PS | PS 0 | 24 (80) |
| PS 1-2 | 4 (13.4) | |
| PS > 2 | 2 (6) | |
| BCLC | Stage 0 | 0 (0) |
| Stage A | 1 (3.8) | |
| Stage B | 19 (73.1) | |
| Stage C | 4 (15.4) | |
| Stage D | 2 (7.7) | |
| Number of focal lesions | Single | 17 (56.7) |
| Multiple | 13 (43.4) | |
| Site of focal lesions | Right lobe | 18 (60) |
| Left lobe | 5 (16.7) | |
| Both | 7 (23.3) | |
| Tumor size by CT | < 3 cm | 1 (3.3) |
| 3-5 cm | 12 (40) | |
| > 5 cm | 17 (56.7) | |
| Portal vein invasion | Yes | 7 (23.3) |
| No | 23 (76.7) |
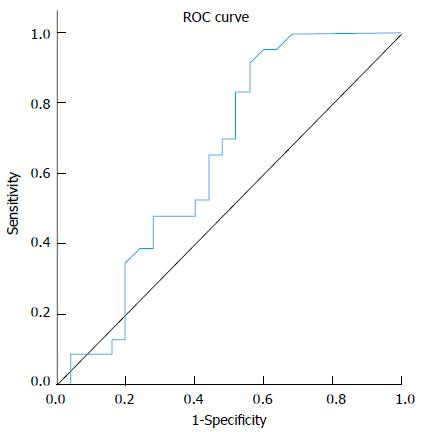
Analysis of median fold change in expression level of miR-122 in patients’ sera in comparison to the normal control group showed that miR-122 displayed significant fold decrease in expression in cirrhosis group (0.8) and significant fold increasing in expression level in both CH (2.1) and HCC (2.15) groups (P≤ 0.01), Table 3. Comparing serum miR-122 expression level between different studied groups displayed an increasing tendency towards statistical significant fold elevation in expression of miR-122 in serum of HCC patients (2.15) in comparison to liver cirrhosis (0.8) with P value 0.083 (AUC = 0.646), Figure 1. No significant fold change in miR-122 expression was found between either (HCC vs CH groups) or (CH vs cirrhosis groups). MiRNA122 showed non-significant up-regulation in HCC patients in comparison to non-HCC patients (CH and Cirrhosis); (P = 0.21).
Analysis of fold change in expression level of miR-221 in patients’ sera in comparison to the normal control group showed significant fold decrease in HCC group and significant fold increase in expression level in CH and cirrhosis groups in comparison to normal control group (< 0.01), Table 4. There was a statistically significant fold decreasing in serum miR-221 levels of HCC patients (0.92) in comparison to cirrhosis (3.4) and in comparison to CH (1.7) (P = 0.05 and 0.06 respectively). On the other hand, there was no statistical significant fold change in serum miR-221 expression level between (CH vs cirrhosis groups) (P = 0.214).
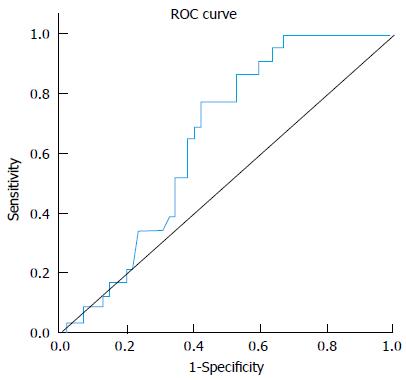
miRNA-221 displayed significant fold decrease in HCC group (0.92) compared to non-HCC patients (CH and cirrhosis) (1.81) (P value 0.03). At a cut-off of 1.82 folds, miR-221 yielded 87% sensitivity and 40% specificity in differentiating between both groups. Figure 2, Table 5.
| AUC | P value | Best cutoff | Sensitivity | Specificity | PPV | NPV |
| 0.655 | 0.03 | < 1.82 | 0.87 | 0.40 | 0.47 | 0.83 |
There was no statistically significant correlation between serum expression level of studied miRNAs and serum AFP level in the different studied groups of patients. No significant correlation was found between the two miRNAs and tumor size, Child-pugh grade in HCC group of patients.
Serum miRNA-122 expression level showed statistically significant correlation with serum necro-inflammatory markers of the liver [aspartate transaminase (AST) and alanine transaminase (ALT) levels) in CH group (P value 0.034 and 0.030 respectively), Table 1.
Over the last 2 decades it has become common practice to use tumour markers, mainly AFP, for the screening of HCC. However, performance of tumor markers has not been optimal with the sensitivity and specificity of AFP and PIVKA-II in the range of 39%-64% and 76%-91%, and 41%-77% and 72%-98%, respectively[9,10], the quest for an optimal tumor marker hence continues. miRNAs have been implicated in roles affecting cellular proliferation and oncogenesis[11]. Cellular miRNAs have been linked with HCC[12]. Their availability in the circulation makes them a tempting target for early tumor detection[12]. The aim of the present study was to explore the potential usefulness of serum miR-122 and miR-221 as novel noninvasive markers for diagnosis of HCV related hepatocellular carcinoma in Egyptian patients.
Most of our HCC patients were within Child-Pugh A and B classifications (56.7%, 36.7% respectively), 73.1% were stage B on BCLC scoring system[13]. This could be explained by the fact that most of them were recruited while being assessed for the possibility of interventional treatment. Another possible explanation for rather good liver condition seen in HCC series could be attributed to that with implementing surveillance programs, allowing detecting tumors at an early stage in well compensated patients. Alfa fetoprotein level was normal (< 10 ng/dL) in 13.4% of recruited HCC patients. Similar finding was observed by Tateishi et al[14] who suggested that not all tumors secrete AFP, and serum levels are normal in up to 40% of small HCCs. It was also showed that α-Fetoprotein alone is not recommended for the diagnosis of HCC and studies showed that its cut off value should be set at 200 ng/mL.
Analysis of fold changes in expression level of miR-122 displayed significant fold increase in expression level in chronic hepatitis C group (2.1) and significant fold decrease in expression in cirrhotics (0.8) in comparison to normal controls. miR-122 is present abundantly in hepatocytes with much lower levels in the circulation in healthy subjects. With hepatocyte injury miR-122 is released in the circulation more readily and serum levels rise. With the eventual loss of hepatocytes and development of fibrosis with proliferation of myelofibroblasts and accumulation of extracellular matrix the circulating miR-122 levels drop again[15].
In the current study there was significant fold rise in serum expression level of miR-122 in HCC group in comparison to normal control group (P value < 0.01). Matching our results, Trebicka et al[15] who studied hepatic miR-122 expression in 43 HCV related HCC in comparison to 3 healthy liver samples using qRT-PCR; miR-122 was strongly up-regulated in malignant liver nodules in comparison to healthy liver. They suggested that miR-122 might down regulate target mRNA of unknown tumor suppressor genes and thus lead to further tumor growth[15].
In a study on hepatitis B patients Xu et al[16] suggested that cancer-induced hepatocyte damage would release the abundant intracellular miR-122 into the circulation, the stability of miRNA would be reflected by easily detectable high blood levels[17]. In contrast to our results, significant down regulation of miR-122 in HCC compared to normal liver tissue was reported by Meng et al[18], Wang et al[19] and Huang et al[20] who compared miR-122 expression profile of 3 different pairs of tumor and normal human liver-derived RNA and 20 HCC liver tissues (mixed etiologies) to normal tissues respectively using microarray[18,19,20]. Similarly a significant down regulation in miR-122 in 19 HBV related HCC liver tissue in comparison to paired healthy liver by next-generation sequencing was reported by Connolly et al[21].
Ladeiro et al[22] have established significant down expression of miR-122 in 28 HCC liver tissues (mixed etiologies) in comparison to 4 healthy liver tissues by qRT-PCR.
In our series, no statistically significant correlation could be verified between serum miR122 expression level and patient characters (age), liver synthetic functions tests (Albumin, bilirubin and PC), or serum AFP level in HCC vs non HCC group (CH and cirrhosis). However, in the chronic hepatitis groups serum miR-122 was correlated with higher AST and ALT levels, further solidifying the theory regarding the initial rise in miR-122 levels due to hepatocyte inflammation and destruction followed by a drop in the levels with the developing fibrosis. Köberle et al[23] also reported significant correlation between serum miR-122 expression level and necro-inflammatory markers (AST, ALT), and Albumin but no significant correlation was found with bilirubin in HCC patients[23].
Perhaps the most significant finding in our study was related to miR-221. Analysis of fold change in expression level of miR-221 in patients’ sera of HCV associated liver disease (CH and cirrhosis) in comparison to normal control group showed significant fold increase in expression level in CH and cirrhosis groups in comparison to normal control group (< 0.01). Also a significant fold decrease in serum miR-221 in HCC group (0.92) in comparison to normal control was noticed. We assumed that with the progression of liver disease from chronic hepatitis to cirrhosis the increased activity of hepatic stellate cells was associated with increase miR-221 expression level, such high level stimulated tumorigenesis and increase level of miR-221 in tissue, but as miR-221 is anti apoptotic so serum miR-221 didn’t show similar increase. In contrast to our results many studies established up regulation of miR-221 in HCC in relation to normal control, e.g.,[18,19,20,24]. However, most these studies assessed tissue miR-221 rather than serum levels .The different results could also be explained by technical variations including sampling methods and freezing and RNA isolation procedures. The etiology of liver disease is also variable in different studies including viral and alcoholic. The stage of the disease is also a source of variation especially that it is still not evident how miRNA expression changes with fibrosis progression. Different studies have also used different control samples for normalization, e.g., non-HCC tissue from the same patient, healthy liver tissue from another subject or patients with the same pathology but not HCC, this is especialy relevant to studies assessing tissue miRNA levels[25].
Similar to what was previously reported by Rong et al[26], we found no statistically significant correlation could be verified between serum miR-221 expression level and patient characters (age), laboratory values (AST, ALT), liver synthetic functions tests (Albumin, bilirubin and PC), or serum a-fetoprotein level in HCC vs non HCC group (CH and cirrhosis) and no statistically significant correlation could be found between the clinic-pathological parameters of hepatic focal lesion, e.g., (number of focal lesions, Child score, biggest diameter of focal lesion BCLC, and portal vein invasion) and miR-221 expression level (P≥ 0.05)[26].
Circulating miR-221 level is significantly up-regulated in the serum of HCV infected patients. It has some value in the differentiation between HCV patients with hepatocellular carcinoma and those without with 87% sensitivity and 40% specificity. It may be able to serve as a promising non-invasive diagnostic marker for HCC. Better results could be obtained if combined with other markers and testing a panel of miRNA’s collectively could ultimately serve as a reliable diagnostic test for HCC.
Hepatocellular carcinoma (HCC), the most common type of liver cancer, is amongst the top three leading causes of cancer-related deaths worldwide with a median survival of only six to eight months. This poor outcome is related to the late detection, with more than two thirds of patients diagnosed at advanced stages of disease. Thus, surveillance of populations at-risk may detect tumors at an early stage when curative interventions can be implemented. The performance of available circulating biomarkers in the screening and diagnostic settings of HCC is sub-optimal.
MiRNAs constitute a large class of genes that encode short RNAs (19-24 nucleotides long), which play key roles in development and differentiation, by the post-transcriptional regulation of protein coding genes. At present, miRNAs have a widely recognized role in human carcinogenesis, including hepatocarcinogenesis, and many experimental evidences indicate that they may act as oncogenes or tumor suppressor genes regulating the expression of crucial protein coding genes. MiRNAs have been proposed as possible novel biomarkers for cancer diagnosis.
In the current study a signature of circulating miRNAs (miR-122 and miR-221) was evaluated. MiR-221 was differentially expressed between patients with HCC and those without (chronic hepatitis and cirrhosis) with lower serum level of miR-221 in former group of patients in comparison to later one. MiR-221 yielded 87% sensitivity and 40% specificity in differentiating between both groups at a cutoff 1.82 folds.
The present study emphasis that circulating miR-221 deserves much attention as potential non invasive biomarkers for HCC in the diagnostic setting.
HCC: Hepatocellular carcinoma; Non HCC: Chronic hepatis C group of patients and patients with liver cirrhosis.
The manuscript entitled “Circulating microRNA, miR-122 and miR221 Signature in Egyptian Patients with Chronic Hepatitis C Related Hepatocellular Carcinoma”. The manuscript is interesting.
P- Reviewer: Hwang KC, Jiang CP, Zou ZM S- Editor: Ji FF L- Editor: A E- Editor: Wu HL
| 1. | Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37 Suppl 8:S4-S66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1370] [Cited by in F6Publishing: 1492] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB. Hepatocellular carcinoma: an epidemiologic view. J Clin Gastroenterol. 2002;35:S72-S78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 3. | Velázquez RF, Rodríguez M, Navascués CA, Linares A, Pérez R, Sotorríos NG, Martínez I, Rodrigo L. Prospective analysis of risk factors for hepatocellular carcinoma in patients with liver cirrhosis. Hepatology. 2003;37:520-527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 311] [Cited by in F6Publishing: 299] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 4. | Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208-1236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4333] [Cited by in F6Publishing: 4404] [Article Influence: 231.8] [Reference Citation Analysis (0)] |
| 5. | El-Attar IA. Cancer databases in the Arab world. Ethn Dis. 2005;15:S1-3-S1-4. [PubMed] [Cited in This Article: ] |
| 6. | El-Nady GM, Ling R, Harrison TJ. Gene expression in HCV-associated hepatocellular carcinoma--upregulation of a gene encoding a protein related to the ubiquitin-conjugating enzyme. Liver Int. 2003;23:329-337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Fattovich G, Giustina G, Sanchez-Tapias J, Quero C, Mas A, Olivotto PG, Solinas A, Almasio P, Hadziyannis S, Degos F. Delayed clearance of serum HBsAg in compensated cirrhosis B: relation to interferon alpha therapy and disease prognosis. European Concerted Action on Viral Hepatitis (EUROHEP). Am J Gastroenterol. 1998;93:896-900. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 137] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 8. | Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, Reddy KR, Harnois D, Llovet JM, Normolle D. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137:110-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 462] [Cited by in F6Publishing: 502] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 9. | Okuda H, Nakanishi T, Takatsu K, Saito A, Hayashi N, Takasaki K, Takenami K, Yamamoto M, Nakano M. Serum levels of des-gamma-carboxy prothrombin measured using the revised enzyme immunoassay kit with increased sensitivity in relation to clinicopathologic features of solitary hepatocellular carcinoma. Cancer. 2000;88:544-549. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 10. | Marrero JA, Lok AS. Newer markers for hepatocellular carcinoma. Gastroenterology. 2004;127:S113-S119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 11. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2105] [Cited by in F6Publishing: 2246] [Article Influence: 132.1] [Reference Citation Analysis (0)] |
| 12. | Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907-1917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3241] [Cited by in F6Publishing: 3212] [Article Influence: 153.0] [Reference Citation Analysis (0)] |
| 13. | Chen DS, Sung JL, Sheu JC, Lai MY, How SW, Hsu HC, Lee CS, Wei TC. Serum alpha-fetoprotein in the early stage of human hepatocellular carcinoma. Gastroenterology. 1984;86:1404-1409. [PubMed] [Cited in This Article: ] |
| 14. | Tateishi R, Yoshida H, Matsuyama Y, Mine N, Kondo Y, Omata M. Diagnostic accuracy of tumor markers for hepatocellular carcinoma: a systematic review. Hepatol Int. 2008;2:17-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 126] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 15. | Trebicka J, Anadol E, Elfimova N, Strack I, Roggendorf M, Viazov S, Wedemeyer I, Drebber U, Rockstroh J, Sauerbruch T. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 2013;58:234-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 16. | Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 410] [Cited by in F6Publishing: 448] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 17. | Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223-1232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 332] [Cited by in F6Publishing: 364] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 18. | Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2044] [Cited by in F6Publishing: 2116] [Article Influence: 124.5] [Reference Citation Analysis (0)] |
| 19. | Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, Tantoso E, Li KB, Ooi LL, Tan P. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205-13215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 315] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 20. | Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, Li W, Bi J, Zhang LJ, Fu Q. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39:786-794. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 127] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 21. | Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, Zavolan M, Tuschl T, Rogler CE. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856-864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 212] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 22. | Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955-1963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 527] [Cited by in F6Publishing: 568] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 23. | Köberle V, Kronenberger B, Pleli T, Trojan J, Imelmann E, Peveling-Oberhag J, Welker MW, Elhendawy M, Zeuzem S, Piiper A. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer. 2013;49:3442-3449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 142] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 24. | Zekri A, Youssef AS, Ahmed O. Serum MicroRNAs as differential markers and a molecular therapeutic target for HCV associated liver disease. AASLD. 2013;poster. [Cited in This Article: ] |
| 25. | Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371-1383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 184] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 26. | Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 98] [Article Influence: 8.9] [Reference Citation Analysis (0)] |