Published online Jan 26, 2021. doi: 10.4252/wjsc.v13.i1.1
Peer-review started: June 26, 2020
First decision: September 18, 2020
Revised: September 28, 2020
Accepted: November 9, 2020
Article in press: November 9, 2020
Published online: January 26, 2021
The recent progress in derivation of pluripotent stem cells (PSCs) from farm animals opens new approaches not only for reproduction, genetic engineering, treatment and conservation of these species, but also for screening novel drugs for their efficacy and toxicity, and modelling of human diseases. Initial attempts to derive PSCs from the inner cell mass of blastocyst stages in farm animals were largely unsuccessful as either the cells survived for only a few passages, or lost their cellular potency; indicating that the protocols which allowed the derivation of murine or human embryonic stem (ES) cells were not sufficient to support the maintenance of ES cells from farm animals. This scenario changed by the innovation of induced pluripotency and by the development of the 3 inhibitor culture conditions to support naïve pluripotency in ES cells from livestock species. However, the long-term culture of livestock PSCs while maintaining the full pluripotency is still challenging, and requires further refinements. Here, we review the current achievements in the derivation of PSCs from farm animals, and discuss the potential application areas.
Core Tip: The successful derivation of pluripotent stem cells (PSCs) from livestock represents an ideal model for the progress of veterinary, biomedical and regenerative medicine. The inherent properties of self-renewal and differentiation make PSCs an ideal raw biomaterial for innovative approaches in artificial reproductive techniques, cell-based therapy, disease modelling, drug testing, organ generation, breed conservation and in vitro meat production. In this review, we present the current status of PSCs application for the development of livestock farming and their potential applications for human welfare.
- Citation: Kumar D, Talluri TR, Selokar NL, Hyder I, Kues WA. Perspectives of pluripotent stem cells in livestock. World J Stem Cells 2021; 13(1): 1-29
- URL: https://www.wjgnet.com/1948-0210/full/v13/i1/1.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i1.1
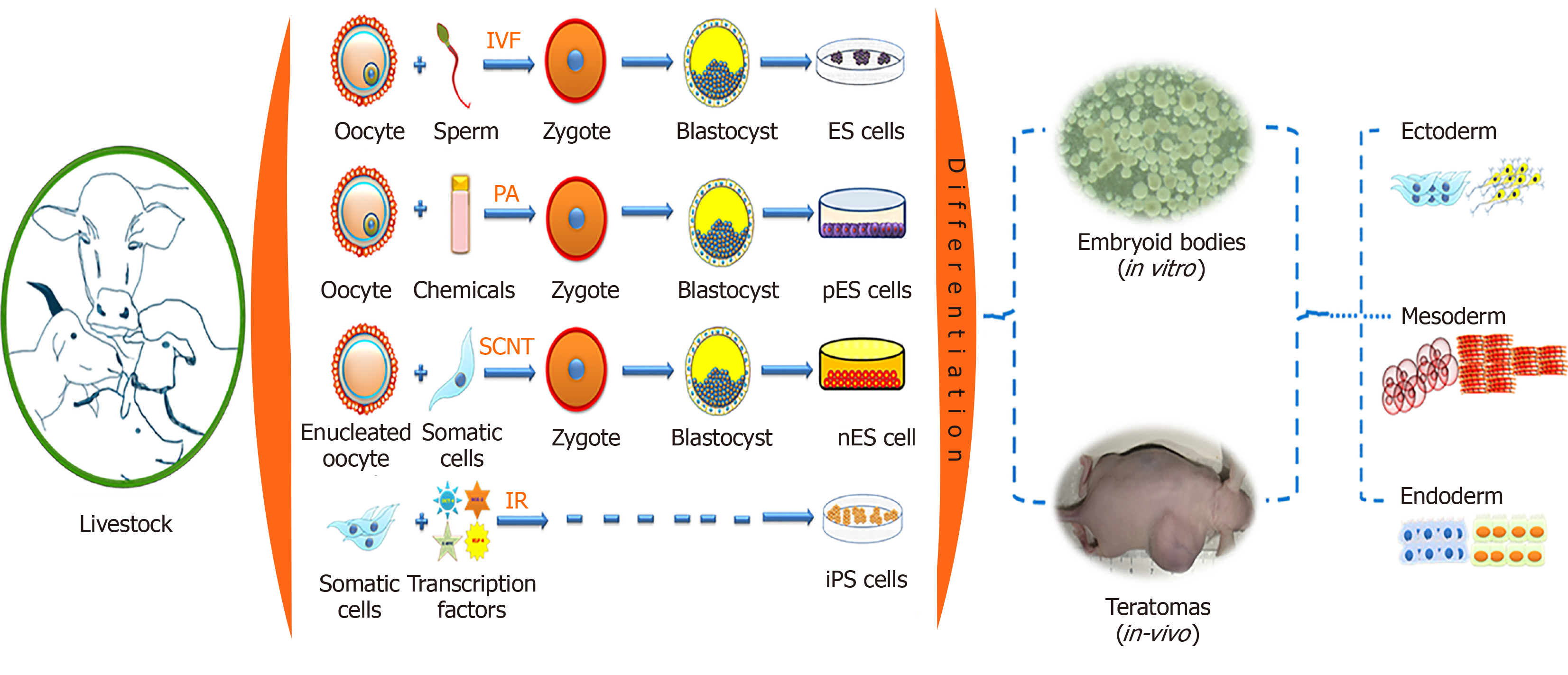
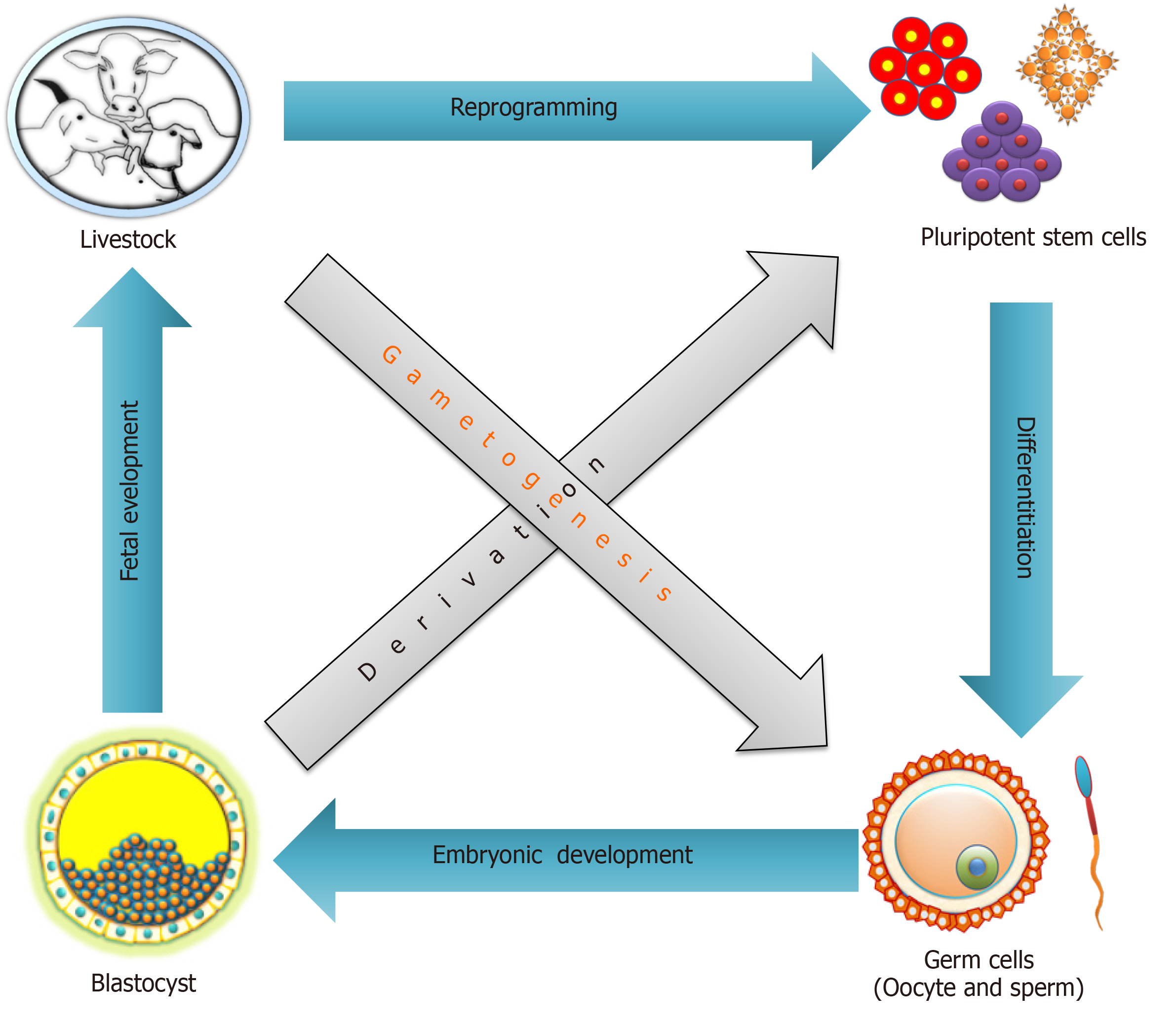
Pluripotent stem cells (PSCs) have the capability to self-renew and to develop into the three primary germ cell layers and therefore can form all cells and tissues of the adult body. There are two sources for obtaining PSCs, embryonic stem (ES) cells developed from an embryo, and induced pluripotent stem (iPS) cells derived via reprogramming of somatic cells (Figure 1). The process of fertilization, parthenogenetic activation, or nuclear transfer (NT), can lead to zygote formation followed by rapid cleavage divisions, which eventually results in the blastocyst stage with two different cell compartments, the outer trophectoderm and the inner cell mass (ICM). After zygote formation, the embryo undergoes several genetic and epigenetic changes, such as DNA de-methylation and re-methylation, replacement of protamines to histones, telomere extension, histone reprogramming, and first activation of the embryonic genome[1]. The resulting ICM cells in the blastocyst have a transient cellular pluripotency and will later form the embryo proper, and thus are able to develop into all somatic cells of an organism. The first successful derivation of cell cultures from the ICM, which maintain these pluripotent properties in vitro, was described in 1981 with murine cells, which were termed ES cells[2,3]. Under specific in vitro conditions, such as the culture on feeder cell layers, the pluripotent status of ES cells becomes locked in the Petri dish. The ES cells showed an unlimited proliferative capacity, were able to be maintained in an undifferentiated state of potency (naïve pluripotency), and could be triggered to differentiate into any cell type. Consequently, ES cells developed into an important arsenal for developmental biology, and new reproduction approaches, such as blastocyst complementation assays and generation of cell chimeric animals, or in vitro differentiation of desired cell types, including gametes[4,5].
However, translation of the protocols for the derivation of ES cells to livestock species is painfully slow. Almost a decade later in 1990, putative ES cells from the early stages of embryos were reported in domestic livestock species such as sheep, pig and cattle; however, these cells could be maintained only for a few passages[6,7]. Later, ES cell-like lines have been derived from many species of livestock such as pig[8,9], cattle[10-13], sheep[14,15], goat[16,17], horse[18], and buffalo[19,20]; however, detailed characteri-zations suggested that these putative ES cell cultures seem to be in a primed status of cellular potency.
NT describes the transplantation of a somatic cell or nucleus in an enucleated oocyte, subsequently, the re-constructed zygote is activated and cultured up to the blastocyst stage. This requires successful reprogramming of the donor nucleus by factors accumulated in the cytoplasm of the recipient oocyte. The NT-derived blastocyst can then be used to derive ES cells from the ICM (‘therapeutic cloning’)[21]. NT-ES cell lines have been established in mice[22-24], cattle[21], buffalo[25] and non-human primates[26]. In livestock, NT-ES cells could be derived from genomically selected high value animals with potential use in reproductive cloning or for conservation using cryopreservation of these cell lines[27].
Alternatively, parthenogenetically derived embryos are equally valuable for the generation of ES cells. The first parthenogenetic embryos derived ES (pES) cell lines were established from mice[28]. Thereafter, it was established in other farm animals such as in pig[29], horse[18], sheep[14], cattle and buffalo[30-33]. Muzaffar et al[32] successfully established buffalo ES cell lines from blastocysts derived from in vitro fertilization, parthenogenesis, and NT. These results suggested that the cell line generated from parthenogenetically derived embryos maintained the ES cell properties and could be used as a model to study the effects of imprinting. However, isolation and characterization of ES cells from livestock species is technically still challenging, as the derived lines showed variable expression patterns of pluripotency markers[34] and may undergo spontaneous differentiation; limited or failed contribution of the transferred ES cells to a chimeric organism in blastocyst complementation assays suggested a limited cellular potency[11,35,36].
In 2008, the 3 inhibitor (3i) approach resulted in the first isolation of authentic rat ES cells[37]. Basically, the 3i approach is an ES culture medium supplemented with three inhibitors of metabolic pathways, which interact with cellular potency: CHIR99021 (GSK3 kinase inhibitor), PD184352 (ERK 1-2 kinases inhibitor), and SU5402 [fibroblast growth factor (FGF) tyrosine kinase receptor inhibitor]. The application of this strategy may allow the generation of genuine PSCs from livestock.
A recent approach is the reprogramming of somatic cells to iPS cells by forced expression of a set of key reprogramming factors such as octamer-binding transcription factor 4 (Oct4), Nanog, MYC proto-oncogene (c-Myc), Kruppel-like factor 4 (Klf4), etc[38-41]. Similar to ES cells, iPS cells are characterized by their self-renewal ability, morphological resemblance, expression of stemness gene, epigenetic state, and their differentiation potential toward all somatic cell types including the germ-line. This technique has been swiftly and widely adopted in farm animal species such as pig[42-47], sheep[48-50], goat[51,52], dog[53-56], cattle[57,58], and horse[59-64]. At present, bona fide ES cells from livestock are not yet available, but long-term stable iPS cells have been generated from many species and allow the further optimization of culture conditions.
The majority of generated livestock iPS cells showed classical features of pluripotency, such as in vivo differentiation and teratoma formation. It has been claimed that porcine iPS cells can contribute to chimera formation[45]. Similarly, ovine iPS cells also formed chimeric lambs after aggregation with early embryonic stages[65-67]; however, the efficiency of chimera contribution warrants further studies. Nevertheless, the results represent advancements in iPS cell technology and promoted the molecular understanding of livestock pluripotency.
PSCs can also be derived from germline stem cells, a class of unipotent stem cells which reside in testis tissue. Observations made by several researchers that neonatal and adult mouse testis contains spermatogonial stem cells (SSCs) or male germline stem cells, which are phenotypically similar to ES cells, are capable of differentiating into three embryonic germ lineages in vitro, form teratomas, and showed germline contribution and transmission[68-71], suggested that the SSCs may retain the ability to generate PSCs. The PSCs derived from these approaches are a useful tool for examining the molecular mechanisms of pluripotency in the male germline. Potentially, the SSCs will be useful for cell-based therapies which benefit males. Similar to SSCs, female germline stem cells (FGSCs) have been proposed to reside in the ovary. Wang et al[72] derived FGSCs from ovaries, which exhibited properties similar to those of ES cells in terms of stemness gene expression and differentiation potential. These novel approaches provide new opportunities to study germ cell biology and opens the possibility of using these cells for genetic diseases in various cell lineages and provides a foundation for personalized regenerative applications. Recent research directed at producing artificial germ cells from stem cells of non-germ line cells may offer the possibility of treating infertility in the future[73].
PSCs derived from ES cells, iPS cells or the germ cell lineage may have great potential in veterinary medicine, cell-based therapies, production of pharmaceutical molecules via transgenic animals, multiplication of elite animals, conservation of endangered animals and as model animals for effective biomedical applications[74-80]. Previously, several review articles on livestock iPS cells have been published which described methods of cellular reprograming and their potential applications[78-83]. Here, we provide the most current achievements in PSCs from livestock, and discuss the potential applications of PSCs for the development of livestock farming and their potential applications for human welfare.
In general, putative livestock ES cells have been derived from early embryonic stages applying either standard culture systems developed for the culture of murine or human ES cells. However, most livestock ES cells did not maintain robust self-renewal and typically failed to perform in teratoma and blastocyst complementation assays[84,85]. This may be due to differences in the ontogenesis of cell lineage formation between rodent and livestock species. In rodent embryonic development, the first cellular differentiation is initiated at the late morula stage where the outer cells develop into an epithelial structure, followed by blastocoel formation. This leads to the development of two cell lineages, the trophectoderm (TE) and ICM[86]. The ICM differentiates further into the epiblast and the primitive endoderm or hypoblast. The epiblast, hypoblast and TE are common in all mammalian blastocysts, but the timing since fertilization in livestock embryos is relatively delayed compared to the mouse[34,87-89].
In porcine and bovine embryos, the hypoblast cells form on 7/8 d post-fertilization, and the epiblast forms on day 12[90,91]. Therefore, the time points used to isolate ES cells are not equivalent between livestock and rodent counterparts[92]. Also differences in the molecular pathways that control pluripotency between murine and domestic animals have been elucidated[93-95].
The current concept of different pluripotency states, which are termed naïve and primed, is likely to provide new approaches for the derivation of livestock ES cells. A steady progress has been made toward optimizing culture conditions for the derivation of stable and highly potent porcine ES cells[8,96]. More recently, porcine ES cells have been claimed to give rise to the chimeric contribution using a modified medium supplemented with basic fibroblast growth factor and leukemia inhibitory factor; however, follow-up studies with respect to contribution to the germline and formation of functional gametes are warranted[97,98]. Similarly, canine ES cells were derived; these canine ES cells expressed all the pluripotent markers, showed long-term self-renewal, and formed teratomas[99]. Thus, these cell lines exhibited most hallmarks of genuine ES cells, which represent a step toward pre-clinical therapies in large animal models.
However, these efforts on establishing livestock ES cells have not been turned into bona fide ES cell lines that are competent in germ line transmission. Multifactorial reasons may contribute to the lack of success for this relevant aim. First, significant differences in the initial embryonic development in livestock from that in rodents, second, the established pluripotency markers may be less distinct for livestock ES cells, and third, the pluripotency states, naïve vs primed, are pretty poorly defined in livestock species[84,100]. Several reviews have been published on the topic of ES cells in livestock covering isolation stages, culture condition, differentiation, proliferation properties and characterization[85,89,92,101-103]. These reviews summarized our knowledge on livestock ES cells. Table 1 summarizes some of the most recent observations that have been reported for ES cells from livestock.
| Species | Embryonic stage | Culture medium and condition | Expression of pluripotency markers | Long-term culture (passage) | Karyotype | In vitro differentiation to EBs | In vivo differentiation to teratoma | Germ line transmission | Ref. |
| Cattle | CDX2-KD blastocysts | KO-DMEM, 2 mM glutamine, 1% MEM-NEAAs, 20 ng/mL hrFGF, 20 ng/mL hrLIF, 0.1 mM β–mercaptoethanol, 15% FBS + MEF under 37℃, 5% CO2 | Yes | 37 | Normal | Yes | Yes | No | [272] |
| Blastocysts | CTFR medium contains low fatty acid BSA, 20 ng/mL hFGF2, 2.5 μM IWR1 + MEFs under 37℃, 5% CO2 | Yes | > 70 | Normal | No | Yes | No | [145] | |
| Buffalo | Blastocysts | KO-DMEM, 15% KSR, 2 mM L-glutamine, 50 μg/mL gentamicin sulfate, 1% MEM-NEAAs, 0.1 mM β-mercaptoethanol, 1000 IU/mL mLIF, 5 ng/mL FGF2 + BFF under 37℃, 5% CO2 | Yes | 135 | Normal | Yes | No | No | [20] |
| Blastocysts | DMEM, 20% FBS, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 2% NEAA, 1% ITS, 50 μg/mL gentamycin sulfate, 30 ng/mL LIF, 40 ng/mL bFGF + BFF under 38.5℃, 5% CO2 | Yes | 15 | Normal | No | Yes | No | [273] | |
| Ovine | Blastocysts | DMEM high glucose, 2 mM L-glutamine, 1 mM Na-Pyruvate, 0.1 mM β-mercaptoethanol, 0.1 mM NEAAs, 10 ng/mL LIF, 20 mg/mL insulin, 1000 IU/mL penicillin, 10 mg/mL streptomycin + STO under 38.5℃, 5% CO2 | Yes | No | Normal | Yes | No | No | [274] |
| Blastocysts | DMEM/F12 supplemented with N2, B27, GSK3 inhibitor (CHIR99021), rhbFGF + OEF or MEF under 38.5℃, 5% CO2 | Yes | 30 | No | Yes | Yes | No | [275] | |
| Caprine | Blastocysts | DMEM, 20% FCS, 1000 IU/mL mLIF, 1% NEAAs 0.1 mM β-mercaptoethanol, 2 mM l-glutamine + GFF under 38.5℃, 5% CO2 | Yes | 15 | Normal | Yes | No | No | [17] |
| Blastocysts | DMEM, 0.1 mM 2-mercaptoethanol, 0.1 mM MEM-NEAAs, 2 mM L-glutamine, 10% FBS, 1000 U/mL hLIF + GFF under 37℃, 5% CO2 | Yes | 120 | Normal | Yes | Yes | No | [16] | |
| Porcine | Blastocysts | 1:1 ratio of 1. α-MEM medium supplemented with 10% KSR, 0.05 mM β-mercaptoethanol, 1% NEAAs, 1% antibiotic-antimycotic, 4 ng/mL EGF, 10 μL/mL 100 × ITS, 1000 U/mL mLIF, 2 ng/mL bFGF and, 2. DMEM/F-10-based medium supplemented with 15% heat-inactivated FBS, 0.2 mM β-mercaptoethanol, 1% NEAA, 1% antibiotic–antimycotic and 2 ng/mL bFGF + MEFs under 37℃, 5% CO2 | Yes | 19 | Normal | Yes | No | No | [276] |
| Blastocysts | α-MEM, 20% KSR, 20 ng/mL bFGF, 20 ng/mL EGF, 10 ng/mL Activin-a, 1% ITS, 1 mM MEM-NEAAs, 55 μM β2-mercaptoethanol + STO at 38.5℃, 5% CO2 | Yes | 21 | Normal | Yes | Yes | No | [277] | |
| Blastocysts | DMEM, 20% KSR and N2B27 medium, 1% NEAAs, 2 mM L-glutamine, 1% PS, 0.1 mM b-mercaptoethanol, 3 mM CHIR99021, 1 mM PD0325901, 2 mM SB, and 50 ng/mL vitamin C + MEFs under 38.5℃, 5% CO2 | Yes | 139 | Normal | Yes | Yes | No | [278] | |
| Equine | Blastocysts | DMEM/F12, 15% FCS, 1000 U/mL hLIF, 15% FBS + MEF under 38.5℃, 5% CO2 | Yes | 28 | Normal | Yes | No | No | [279] |
| Blastocysts | KO-DMEM, 15% FBS, 0.1 mM NEAAs, 2 mM L-glutamine, 1% ITS, 100 μg/mL streptomycin, 100 IU/mL penicillin, 0.1 mM β-mercaptoethanol, hLIF, hbFGF + MEF under 38.5℃, 5% CO2 | Yes | 15 | No | No | No | No | [280] | |
| Canine | Blastocysts | KO-DMEM/Ham’s F12, 15% KSR, 1 × GlutaMAX, 1 × NEAAs, R3IGF1, 0.1 mM 2-mercaptoethanol, 10 ng/mL hrLIF, 4 ng/mL rhFGF2, 0.5 μM, MEK inhibitor PD0325901, 3 μM GSK3β inhibitor CHIR99021 + MEFs under 37℃, 5% CO2 | Yes | -- | Normal | Yes | Yes | No | [281] |
| Blastocysts | KO-DMEM or DMEM/-12, 0.1 mM β-mercaptoethanol, 5 μM thymidine, 15 μM cytidine, 15 μM guanosine, 15 μM adenosine and 15 μM uridine nucleosides, 0.2 mM GlutaMax, 0.1 mM NEAAs, penicillin (100 IU/mL), streptomycin (50 μg/mL), 10 ng/mL hLIF, 4 ng/mL hbFGF, 15% FBS or KSR + MEFs under 37.5℃, 5% CO2 | Yes | 30 | Normal | Yes | Yes | No | [99] |
A ray of hope was the development of reprogramming techniques to obtain iPS cells from somatic cells[38]. Recent advances in iPS cells technology may overcome the bottleneck of establishing pluripotent cells from livestock species, and demonstrate that iPS cells showed advanced level of pluripotency, such as the ability to differentiate in vitro into multi lineages and in vivo into teratomas and chimeras[79,104]. The production of chimeric livestock from iPS cells would open the possibility to genetically engineer farm animals to improve traits of agricultural importance, and the generation of biomedical models[45,67,105].
The current knowledge on stemness gene regulation in ES cells helps to execute iPS cells in a better way, and could allow the development of approaches closer to clinical application. Transcriptional profiling of ES cells revealed that factors such as Oct4, Nanog, sex determining region Y-box 2 (Sox2), Klf4, c-Myc and Lin28 are essential to maintain pluripotency[106,107]. Among these factors, Oct4, Nanog and Sox2 have been identified as core transcription factors, showing both spatial and temporal expression in cultured PSCs and pluripotent cells of the ICM[108-110]. These core transcription factors also play pivotal roles in regulation of the pluripotent gene expression and simultaneous suppression of many genes related to differentiation[111,112]. They exert their functions by co-occupying their target genes, and can bind at their own and each other’s promoters to form an interconnected auto-regulatory loop[111,113]. These three factors function collaboratively in an auto-regulatory circuitry fashion to maintain their own expression and maintain the pluripotency of ES cells[106,114]. Additionally, Nanog was identified as the key factor, which regulates the establishment of the pluripotent epigenome[115,116]. The role and mechanism of these transcription factors in the reprogramming of livestock somatic cells to iPS cells have been reviewed[78,81,104,117].
Several strategies have been applied to deliver core reprogramming factors such as genes, mRNAs and proteins into somatic cells for the derivation of iPS cells. Alternatively, the replacement of reprogramming factors by small chemical agents has also been assessed. Commonly, retro- and lenti-viral approaches were employed for cellular reprogramming of different types of somatic cells from livestock such as pig[105], cattle[57,118-120], sheep[67], goat[51,121], dog[54,56], horse[63] and buffalo[65]. Recently, for the first time, cat iPS cells were created using disarmed retroviruses with the coding sequences for human Oct4, Sox2, Klf4, cMyc, and Nanog[122]. The expression of ectopic factors can be temporally confined by employing inducible promoters or viral promoters, which are epigenetically silenced. Shortcomings of the viral approach include the limited cargo capacity of approximately 7 kb for the transduced genes, the induction of innate immune responses, potential genotoxic effects, which limit the translation into clinical trials[123], and increased safety methods.
To evade these safety concerns, remarkable technological progress has resulted in the establishment of non-integrating viral and non-viral approaches, but limited attempts have been made to apply these to cells from livestock species. For example, the non-integrating adeno- and Sendai-viruses demonstrated efficient production of human, murine and canine iPS cells[124-127]. Apart from viral-mediated derivation of livestock iPS cells, non-viral approaches such as plasmid vectors, recombinant proteins, transposons, minicircle DNAs, small molecules, and mRNAs are in use to eliminate the risk of genomic alteration and enhance the prospects of iPS cells. In this regard, bovine iPS cells were successfully derived by plasmid[128], and transposon systems[58,129,130]. Similarly, porcine iPS cells were also established using episomal[131,132], and transposon systems[47], the transposon systems have been attempted to derive equine[59] and buffalo[66] iPS cells. Detailed information on approaches to generate transgene-free iPS cells has recently been reviewed by Haridhasapavalan et al[82]. Most recent studies of iPS cells derived from livestock are shown in Table 2. In addition, extensive overviews of iPS cells produced from a wide range of animal species including livestock with their prospective applications and limitations have been recently reviewed[85,103,133]. Most recently, our group presented the potential applications of transposon-mediated derivation of iPS cells for cell-based therapies[83].
| Species | Cell type | Culture medium and condition | Expression of pluripotency markers | Long-term culture (passage) | Karyotype | In vitro differentiation to EBs | In vivo differentiation to teratomas | Germline transmission | Ref. |
| Cattle | Fetal fibroblasts | DMEM/F-12, 20% KSR, 1 mM l-glutamine, 0.1 mM NEAAs, 0.1 mM mercaptoethanol, 100 U/mL penicillin, 100 μg/mL streptomycin, 8 ng/mL bFGF, 1000 U/mL hLIF on MEF at 37℃ and 5% CO2 | Yes | 40 | Normal | Yes | Yes | No | [58] |
| Fetal fibroblasts | KO-DMEM, 15% FBS, 2 mM L-glutamine, 1% NEAAs, 0.1 mM β-mercaptoethanol, 106 U/mL hLIF, 10 ng/mL bFGF on STO at 37℃ and 5% CO2 | Yes | 50 | Normal | Yes | Yes | No | [130] | |
| Buffalo | Fetal fibroblasts | DMEM high glucose, 20% ESC-FBS, 2 mM l-glutamine, 1% NEAAs, 0.1 mM β-mercaptoethanol, 10 ng/mL bFGF, 10 ng/mL LIF on MEF at 37℃ and 5% CO2 | Yes | 10 | Normal | Yes | Yes | No | [65] |
| Fetal fibroblasts | DMEM/F-12, 20% KSR, 0.1 mM NEAAs, 1 mM L-glutamine, 0.1 mM mercaptoethanol, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 ng/mL bFGF, 1000 U/mL hLIF on gelatine at 37℃ and 5% CO2 | Yes | 15 | Normal | Yes | No | No | [66] | |
| Ovine | Embryonic fibroblasts | DMEM, 20% FBS, 1% ITS, 0.1 mM 2-β mercaptoethanol, 1 mM NEAAs, 2 mM glutamine, 4 ng/mL bFGF, 1000 U/mL mLIF on MEFs at 37℃ and 5% CO2 | Yes | 17 | Normal | Yes | Yes | Yes (formation of ICM in tetraploid) | [50] |
| Embryonic fibroblasts | KO-DMEM, KSR, 0.1 mM NEAAs, 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol, 8 ng/mL hFGF2, 1000 U/mL mLIF on SNL at 37℃ and 5% CO2 | Yes | 23 | Normal | Yes | Yes | Yes (live-born chimeric lambs) | [67] | |
| Caprine | Fetal fibroblasts | DMEM/F12, 20% KSR, 1 mM L-glutamine, 0.1 mM 2-mercaptoethanol, 1% NEAAs, 2% sodium bicarbonate solution, 1000 IU/mL 2i/LIF, 4 ng/mL bFGF on STO at 37℃ and 5% CO2 | Yes | 30 | Normal | Yes | Yes | No | [282] |
| Embryonic fibroblasts | KO-DMEM, 20% KSR, 1% NEAA, 1% L-glutamine, 0.1 mM EAA, 1% penicillin /streptomycin, 10 ng/mL FGF2 on GEF at 37℃ and 5% CO2 | Yes | 22 | Normal | Yes | No | No | [283] | |
| Porcine | Embryonic fibroblasts and microvascular pericyte cells | LCDMV medium contains 50% neurobasal medium, 50% DMEM/F12, 1 × N2, 0.5 × B27, 5% KSR, 10 ng/mL LIF, 1 μM CHIR99021, 2 μM (S)-(+)-dimethindene maleate, 2 μM minocycline hydrochloride, 40 μg/mL vitamin C on MEF at 37℃ and 5% CO2 | Yes | 28 | Normal | Yes | Yes | Yes (chimeric formation in post-implantation pig conceptuses) | [284] |
| Sertoli cells | DMEM/F12, 10% KSR, 10% FBS, 1 mM l-glutamine, 1 mM antibiotic, 1% NEAAs, 0.1 mM β-mercaptoethanol, 10 ng/mL bFGF, 10 ng/mL hLIF on MEF at 37℃ and 5% CO2 | Yes | 50 | Normal | Yes | Yes | No | [285] | |
| Equine | Fetal fibroblasts | DMEM/F12, 20% KSR, 10 ng/mL bFGF, 1% penicillin/streptomycin, 10 ng/mL hLIF on MEF at 37℃ and 5% CO2 | Yes | 25 | -- | Yes | No | No | [286] |
| Fetal fibroblasts | DMEM, 20% FBS or KO-DMEM, 20% KSR, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol, 0.1 mM MEM- NEAAs, 1% penicillin–streptomycin, 8 ng/mL hbFGF, 1000 U/mL hLIF on SNL at 37℃ and 5% CO2 | Yes | 30 | Normal | Yes | Yes | No | [61] | |
| Canine | Embryonic fibroblasts | Serum-free N2B27-based medium, 4 ng/mL hbFGF on MEF at 37℃ and 5% CO2 | Yes | 50 | Normal | Yes | No | No | [287] |
| Fetal fibroblasts | KO-DMEM/F12, 20% KSR, 2 mM L-glutamine, 0.1 mM NEAAs, 0.1 mM β-mercaptoethanol, 0.1 mM bFGF on MEF at 37℃ and 5% CO2 | Yes | 15 | Normal | Yes | Yes | No | [288] |
A long-standing goal of PSCs research is the differentiation into functional germ cells, and their application for in vitro fertilization to obtain sexually recombined genotypes. In combination with the readout of genomic trait values from few cells via single nucleotide polymorphism chips and whole genome sequencing techniques this will dramatically improve the breeding process[134]. In mammals, germ cells originate from PGCs, the PGCs are initially specified outside the post-implantation embryo through gradients of Wnt family member 3 (WNT3) and bone morphogenetic proteins (BMPs)[135-137]. After that PGCs migrate to the genital ridges, where they settle and ultimately make gametes. The PSCs are principally immortal, with a high proliferative rate, and the ability to differentiate into gametes that could enable in vitro breeding schemes for accelerated genetic improvement in livestock. Under the conventional breeding scheme of dairy animals, the generation interval for sire(s) or dam(s) of bulls is approximately ± 5 years. This time period could be considerably reduced to about 2.5 years using genomic selection approaches[134,138]. Recently, a proposed parental embryos to offspring embryos breeding system will require approximately 2 mo to finish one round of selection, and annual genetic gain will increase approximately 10-fold as compared to the standard genomic selection in dairy animals[134,139]. However, the feasibility of applying this approach to farm animals needs to be proven in field studies.
So far the proof of principle has been provided in rodents, where sperm and oocytes were generated by the differentiation of male or female PSCs[140,141]. These works support the notion that this approach might be translatable to farm animals. However, before the proposed breeding system can be applied in farm animals a series of obstacles need to be overcome. Earlier, it was observed that monkey ES cells could differentiate into primordial germ cell-like cells (PGCLCs) and their differentiation ability was further improved by supplementing a conditioned medium from testicular or ovarian cells with recombinant BMP4, retinoic acid (RA), or stem cell factor[142,143]. Another study showed the possibility of spermatogonial stem cell transplantation in a non-human primate infertility model[144]. The findings laid the basis for the development of future germ cell regeneration in livestock. Recently, the derivation of stable bovine ES cells was reported, which could offer a technical basis for the auxiliary establishment of in vitro germ cell induction in farm animals[145]. Porcine iPS cells have been successfully differentiated into PGCLCs, and xenotransplantation of these cells into the testes of infertile immune-deficient mice resulted in immunohistochemically identifiable germ cells[146,147]. Another study revealed that porcine PGCs could be derived from the posterior pre-primitive-streak epiblast by upregulation of SOX17 and B-lymphocyte-induced maturation protein 1 through activation of WNT and BMP signaling pathways[148]. A number of comprehensive studies dedicated to bovine germ cell differentiation suggested that RA and/or BMPs are important for induction of PSCs[149]. The significance of RA in gametogenesis and meiosis induction has also been reported in buffalo[150,151].
Apart from reduction of the generational interval using in vitro breeding in livestock, the idea of generating gametes in vitro may translate to treatments of infertility, understanding the complexity of gametogenesis, and it could also be a source for regenerative medicine[78,152]. Improvements in germ cells differentiation of PSCs from livestock will further fuel the enthusiasm of researchers working on farm animals (Figure 2). If robust and field-applicable protocols for in vitro germ cell differentiation of livestock PSCs could be developed, an in vitro breeding program will rapidly be implemented.
More recently, haploid stem cells (hSC), having a single set of chromosomes, are considered excellent tools to study gene function (due to having a single copy) and obviate the mutation effect[153]. To date, hSCs have been derived from mouse, rat, monkey and humans[153,154]. In nature, ova and sperm are haploid cells. Experimentally it has been shown that it is possible to generate murine hSCs containing only the maternal genome or the paternal genome through either parthenogenetic or androgenetic embryos. Recently, it was demonstrated that fertile adult mice can be produced after fertilization of a sperm with an ovum derived from haploid ES cells[155], supporting the significance of haploid stem cells as a new tool to quickly generate genetic models for the direct transmission of genomic modifications at the organism level.
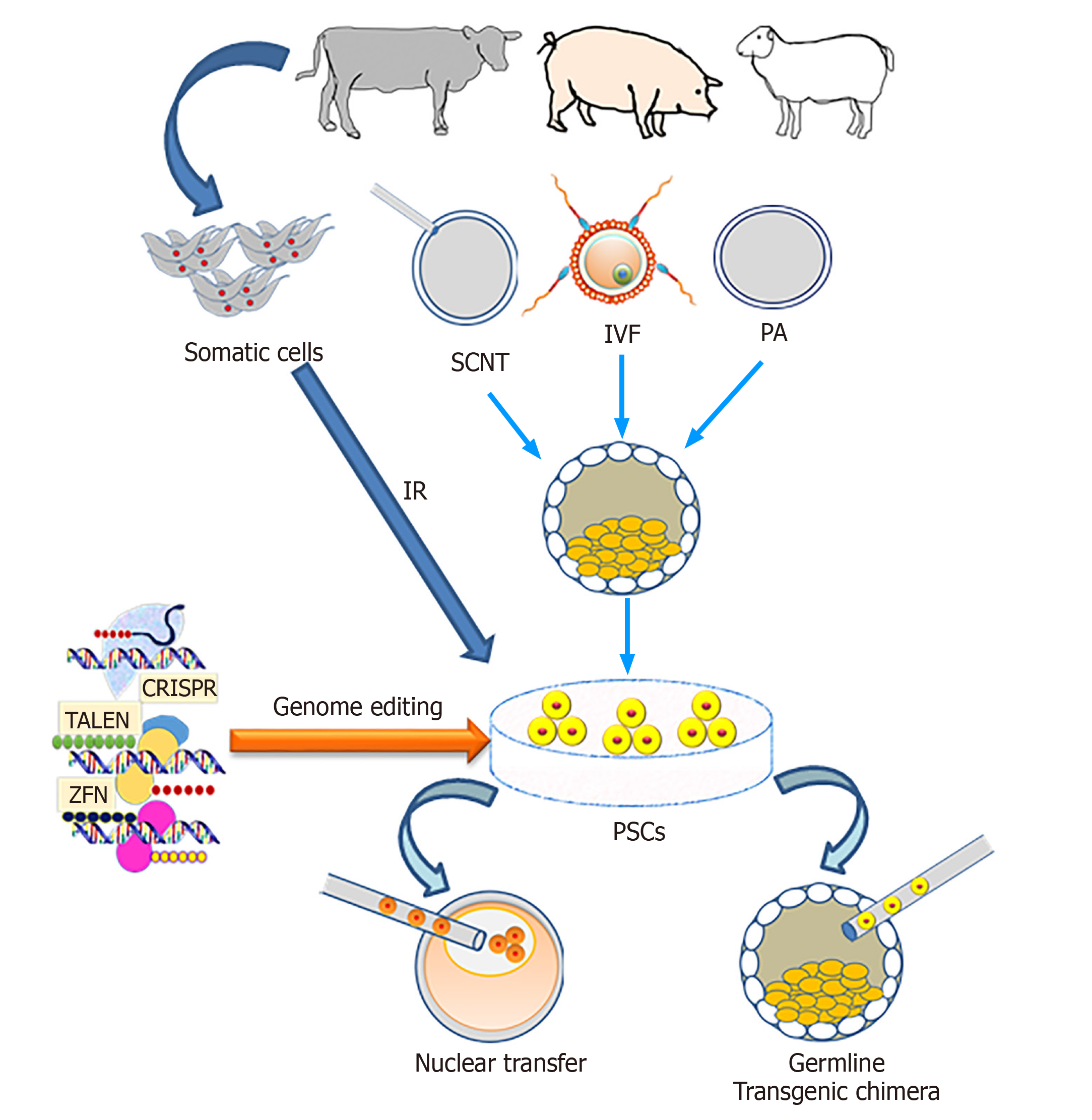
The genetic engineering of animals refers to adding, changing or removing certain DNA sequences, and the inheritance of these modifications to the next generation. The self-renewal and differentiation ability of PSCs make these cells an attractive tool for genetic engineering for various downstream applications. The self-renewal property of PSCs means they are theoretically immortal in vitro through symmetric cell divisions, which could provide a possibility for genetic modification and screening of cells, carrying the intended gene modifications.
Usually, genetic modification requires several generations and a large number of animals that could be overcome using PSCs especially in livestock via contribution to the germline[156]. The PSCs can be genetically modified in vitro and then injected into an embryo where they contribute to the germline, resulting in transgenic offspring (Figure 3) and thus reducing the required number of animals to produce the line founders[157].
In murine ES cell-based targeted mutagenesis, homologous recombination (HR) approaches allow the loss-of-function, gain-of-function experiments of desired loci, but also more complex genetic modifications, such as large genetic recombinations, as well as spatial and temporal expression patterns[158-160]. Using this technology, mouse ES cells can be screened and single colony-derived cells produced to employ for blastocyst complementation. However, in livestock due to the lack of bona fide ES cells, the efficiency of HR events is very low. Richt et al[161] produced cattle with knock-out of the prion gene using somatic cell nuclear transfer (SCNT) combined with HR in somatic cells. Details of several genetic engineering methods used for successful creation of modified PSCs including ES and iPS were reviewed[162].
Previously, many workers reported that the use of ES cells as donor nuclei in SCNT resulted in higher birth rates as compared to differentiated somatic cells[25,163], which may be due to the un-differentiated state of PSCs. An advantage of this approach is that homozygous transgenic founders are produced in one generation. The use of iPS cells derived from livestock to produce offspring by SCNT has so far resulted in low success rates. Attempts to clone pigs and sheep using iPS cells resulted in very low efficiencies[121,164,165], whereas in murine experiments cloned animals were produced with similar efficiency using iPS or ES cells[163,166].
It was assumed that the forced expression of exogenous factors in iPS cells may hamper the nuclear reprogramming of donor cells during SCNT[121]. For this reason the possible applications of PSCs for generating genetically modified livestock have been limited. The recent discovery of site-specific nucleases such as zinc finger nucleases, transcription activator-like effector nucleases and CRISPR-Cas9 has allowed us to overcome the bottlenecks of genetic engineering in livestock[167-171]. The detailed description of site-specific nucleases is beyond the scope of the current manuscript, and interested readers can find excellent reviews elsewhere[156,172].
Blood stem cell transplantation is a well-established clinical treatment of leukemias[173]. Nowadays, the feasibility and translation of innovative cell therapies, based on PSCs, is actively studied. In general, cell therapy is the transfer of cells into a patient to heal lesions or cure a disease, which cannot be addressed adequately by existing pharmaceutical interventions. For this purpose, cells may originate either from the patient (autologous cells) or a donor (allogeneic or heterogenic cells). The cells used in cell therapy should have the capability to proliferate in vitro, and to differentiate into specific cell types in a patient. In the last few years, PSCs have received considerable attention for innovative cell therapies, and has resulted in significant progress in the understanding of their characteristics and therapeutic potential in different lineages. PSCs are considered ideal candidates for cell therapy to achieve tissue repair, or to restore and replace diseased cells.
Using PSCs a large number of animal models has already been treated for numerous diseases to assess the effectiveness of innovative cell-therapies[174-176]. The use of ES cells in cell-therapies is limited due to difficulties in patient-specific derivation, immune-rejection and ethical considerations, whereas derivation of iPS cells has overcome these concerns. Before the clinical application of iPS cell-based therapies is approved, they should be properly assessed using appropriate simulated animal models. Traditionally, laboratory animals (rodents) are used as models due to available knock-out or knock-in gene mutants which demonstrate disease phenotypes. Rodent models for cell-therapies do not always accurately mimic the genetically heterogeneous human situation[177,178]. The use of large animal models seems to be more suitable to analyze efficacies and risks in longitudinal pre-clinical tests and regenerative studies using cell-based-therapy[78,179,180]. Large animal models more closely match human patients in terms of life-span, metabolism, physiology, pathophysiology and biomechanics[181-184]. Large animal models will also permit determination of the effective cell dose, to track the fate of transplanted cells, and to assess their functional integration in the host organ[185]. In addition, large animal models also offer comparative models for research due to naturally occurring diseases, such as cancer[186,187].
Among livestock, the pig is considered a suitable animal model for pre-clinical evaluation of the efficacy and safety of novel cell therapies[179,188,189]. For example, porcine iPS cells can be differentiated in vitro into cells of the rod photoreceptor lineage, which were capable of integration into the retina, and generated outer segment-like projections[186,190]. Similarly, improvement of cardiac functions was documented in pig, but also in sheep, dog and rabbit models by assessing the efficacy of cell transplantations. The transplanted cells include skeletal myoblasts, bone marrow cells, cardiac stem cells, and endothelial stem cells[191-193]. Porcine models have also been used for the transplantation of human iPS cell-derived cardiovascular cells for the treatment of acute myocardial infarction, in which improvements were observed in myocardial wall stress, and contractile performance[194]. In addition, porcine iPS cell–derived endothelial cells were transplanted into a murine myocardial infarction model; the results showed an improved myocardial function by paracrine activation[195]. van der Spoel et al[196] analyzed the published reports of pre-clinical studies involving large animals for ischemic heart disease cell therapies and they concluded that large animal models allow prediction of the outcome of clinical trials for efficacy and safety. Hence, large animal models are useful targets for assessing the potential of iPS cell therapies to treat diseases, which are caused by the degeneration of specific cell populations, such as Alzheimer’s disease, Huntington´s disease (HD), spinal muscular atrophy, retinitis pigmentosa, and diabetes[80,186].
It is well established that PSCs (including ES cells and iPS cells) are able to differentiate into any cell type in vivo or in vitro under suitable conditions, whereas the differentiation and reprogramming potentials of some adult stem cells are still under investigation[197]. This indicates that cells have the potential to switch from one cell type to another under the expression of some pluripotent related genes. Recently, progress made in cellular reprogramming and transdifferentiation suggests that it will be possible to generate cells from autologous sources without immunologic rejection and ethical consideration for therapeutic and regeneration purposes[197].
The objective of animal conservation is to maintain biodiversity because elimination of even a single species can interfere with the functioning of an ecosystem[198,199]. The protection of viable populations in their natural habitat (in situ) is one of the best methods for biodiversity conservation. In situ conservation allows the propagation of a small population using multidisciplinary approaches including genetic and ecological characterizations, but is sometimes insufficient for maintaining adequate genetic diversity[200]. Ex situ conservation approaches have been adopted with the aim of establishing viable populations through cryopreservation of animal genetic resources such as sperm, oocytes, somatic cells, and tissues of valuable domestic breeds and for conservation of endangered wild species. Earlier efforts in wildlife cryo-conservation were generally focused on spermatozoa and embryos[201,202]. More recently, somatic cell bio-banking has emerged as an attractive option for cryo-conservation of endangered and valuable farm animal breeds aiming to revive those in the future using assisted reproduction technologies[203-205]. Advancements made in nuclear transfer and stem cell technologies, and the ability to reprogram differentiated somatic cells into embryonic or germ cell lineages prompted the interest in storing somatic cells for offspring production in future[206-208]. Further advancements in cryobiology may make it possible to cryopreserve different types of cells including somatic cells. Among somatic cells, fibroblast cells are preferable due to abundant availability in skin-tissue and easy establishment in cell culture[203,205]. Primary fibroblast cells derived from livestock such as cattle, buffalo, sheep, goat, and pig have been successfully cryopreserved and are being used for various purposes, including SCNT[205,209-212].
However, the efficiency rates of SCNT in many domestic and wild animals are low, and cloning experiments present a bottleneck in this approach. In mouse cloning, the use of pluripotent blastomeres as donor cells has significantly improved cloning efficiency and decreased the incidence of developmental abnormalities[213]. For endangered species, the availability of oocytes and embryos is often restricted, whereas the generation of iPS cells from somatic cells offers a more practical source of stem cells with less moral and ethical restrictions[214]. The morphology of iPS cells from wild and valuable domestic animals resemble those of ES cells[74,215]. The derivation of iPS cells from skin fibroblasts of endangered primate, silver-maned drill and white rhinoceros[74], snow leopard[215], orangutan[216] and endangered felids such as Bengal tiger, serval and jaguar[217], indicate the feasibility of derivation of iPS cells from threatened species. The generated iPS cells could be expanded for banking as a genetic resource, or used in the animal cloning process to produce viable offspring. Alternatively, iPS cells could be differentiated to derive mature and functional oocytes and spermatozoa, which might be used for in vitro fertilization to produce offspring. Furthermore, the availability of iPS cells from diverse species would help to accelerate research progress on evaluating phylogeographic structure, paternity determination, delineating subspecies, assessing gene flow and genetic variation related information, which could be critical for decision-making in managing both ex situ and in situ wildlife populations[218]. More recently, Hildebrandt et al[219] successfully generated ES cells and embryos from the critically endangered northern white rhinoceros. These achievements strengthen the beliefs that modern biotechnologies or iPS cell techniques in collaboration with cloning technology will allow the generation of more offspring from selected parents to ensure genetic diversity and may reduce the interval between generations. In future, the advancements of reproductive techniques and the new knowledge can only be employed when cryopreserved raw biomaterials (germ cells/somatic cells) are maintained, otherwise these would be lost forever[214].
Human medicine requires animal models to test any new drug, as in vitro systems are still not able to model the pathophysiology of a whole organism. However, many preclinical studies of new therapies conducted on rodents and non-human primates failed due to the fact that they do not allow the prediction of safety and effectiveness in human patients[178,186]. For example, rodent models fail to simulate the basic physiological functions of heart diseases due to their faster heart rate. In contrast, large animals are more similar to humans with regard to their life span, physiology, metabolism, and pathophysiology[179,181,183,220]. The generation of PSCs from livestock[45,58,98] is economically valuable and critically important for the establishment of disease models, testing of new drugs and for the production of medically useful substances such as enzymes and growth hormones[221]. Additionally, animal disease models and animal iPS cells allow the establishment of informative assays to test the efficacy of new compounds, their toxicity, and dosing[222].
Previously, it was demonstrated that iPS cells can be differentiated into lineages of cardiomyocytes and hepatocytes, which were used for disease modelling and drug screening[223-225]. Presently, several biotechnological tools are available to generate disease models using either ES cells or iPS cells, creating new possibilities for their use in drug testing[226,227]. In many cases, somatic cells are also exploited for drug testing and validation, thus promoting new drug discoveries. In addition, the validation of existing drugs in new iPS cell models is performed. The availability of patient-specific iPS cells is crucial to discover new personalized therapeutics.
Large animals such as the domesticated pig have been recognized to be important models for studying colorectal cancer, cardiovascular diseases, cystic fibrosis, diabetes, osteosarcoma, Duchenne muscular dystrophy and Alzheimer’s disease[228]. Similarly, cattle have become a relevant model for studying human female fertility vis-a-vis the effects of ageing on fertility[229], uterine infection[230], and ovarian function[231]. In addition, dogs have natural occurring genetic diseases such as hemophilia B[232]. For other diseases that do not occur spontaneously in animals, transgenic animals have been generated such as monkeys with HD and cystic fibrosis-diseased pigs[233]. Induced PS cells generated from HD monkeys were differentiated into neuronal cells in vitro, and showed typical HD-like features; thus, representing an attractive model for investigating HD pathogenesis and therapy[234,235]. Recently, CRISPR-Cas9 was used to generate large animal models of neurodegenerative diseases that can more realistically mimic human disease progression[236]. The derivation of iPS cells from patients with congenital heart disease, and differentiation of these cells into cardiomyocytes has been anticipated to serve as a model system to study disease pathogenesis and for drug discovery[225,237].
A chimera is a composite organism that is composed of at least two genetically different cell populations[238,239]. It can be produced by combining blastomeres from a minimum of two individual embryos, by aggregating two or more sectioned embryos, or by injecting PSC cells into a blastocyst[240]. Tarkowski et al[241] were the first to demonstrate that the aggregation of two sectioned mouse embryos after transfer into the uterus of a surrogate could result in the development of healthy and fertile chimeric animals. If the cells used for chimera generation have differentiation potency they can contribute to form chimera and chimerism rates depend on the potency of the cells. Presently, ES and iPS cells are preferred for aggregation or injection into early embryos due to their pluripotency and ability to contribute to multiple organs of the resulting chimera[5,242-244].
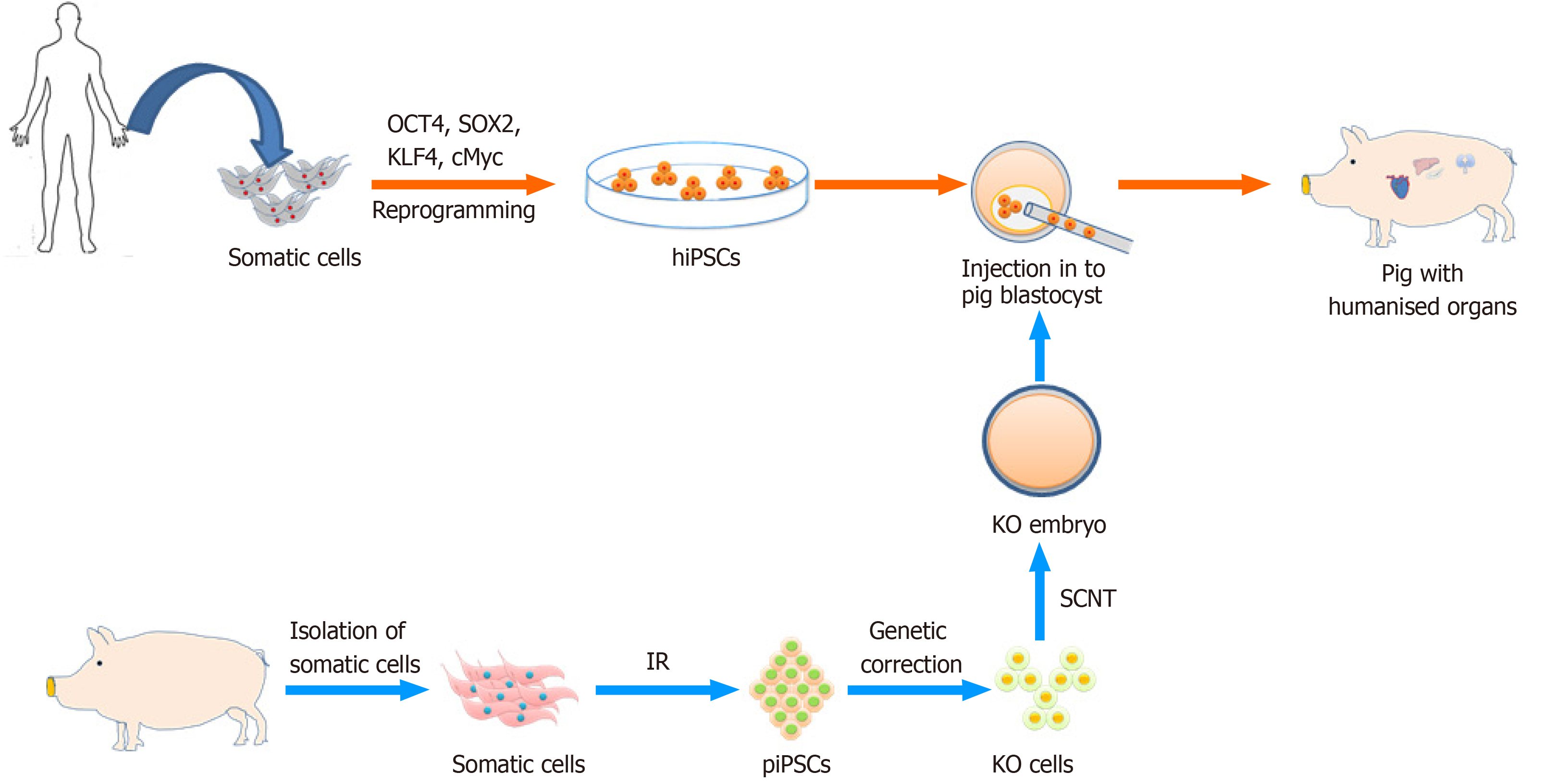
Concurrently, scarcity and demand for human organ donors have motivated scientists to examine options other than donation from deceased patients, such as the possibility of growing human organs in animals. The availability of human PSCs suggested the possibility of producing human organs in animals via the chimera route (Figure 4). As proof of principle it was demonstrated that the combination of mouse/rat cells resulted in viable chimeras, with the possibility to direct the one cell type into an organ-specific lineage. To achieve this one species carries a defective gene (Pdx1) necessary for pancreas development. By complementing the cells with the defective Pdx1 gene, with fully competent PSCs from the other rodent species, a chimera with a functional pancreas may develop. A pancreas formed from rat PSCs has been observed in a mouse host, and a pancreas formed from mouse PSCs in a rat host[242,244]. Interestingly, a rat-sized pancreas formed from mouse PSCs, suggested that it could be possible to produce human organs (xenogeneic in nature) in various animal species[244].
Earlier, rat-mouse[242], human-mouse[245], and sheep-goat[246] chimeras were documented. However, blastocyst injection has failed to introduce stem cells into primate embryos[247]. Aggregation of rat-mouse[248], sheep-goat[249], and cattle-buffalo[250] embryos were able to form interspecies chimeras. In spite of the lower survival rate of chimeric embryos produced by the aggregation method compared with blastocyst injection, the chimerism rates have been observed to be higher[251]. Considering the higher rates of chimerism, the human-animal chimeras could be an organ resource, aggregation is also a desirable choice when the embryo and stem cells are in a good growth condition.
The generation of human organs in animal models would have a significant impact in the field of regenerative medicine, since the shortage of donor organs is a major bottleneck. For the generation of human organs, human PSCs would be injected into blastocysts acquired from carrier animals that should be genetically modified to block the development of a particular organ (Figure 4). Thus, only human cells might predominantly contribute to the development of that organ[76]. Previously, human iPS cells were employed to created chimeras upon integration in porcine and bovine blastocysts[5,244]; however, with very limited colonization of early fetuses by the human cells. Yang created human-mouse chimeras overexpressing functional human coagulation factor IX that could be a suitable candidate for hemophilia B treatment[252]. The feasibility of growing complete human organs using a chimeric approach and the proposed immune tolerance (if autologous donor cells are used) still needs to be substantiated.
More recently, the advent of precision genome editing tools like CRISPR/Cas9, efficiently generating the mutation that leads to organ deficiencies in larger animal models further widening the possibility to create human-animal chimeras[253]. The CRISPR/Cas9 mediated zygote genome editing, already successfully documented for mouse and larger livestock species[254], will likely be an effective tool for chimera production[5]. These results open up horizons in the field of regenerative and personalized medicine, but also impose big challenges of ethical concern with low efficiency of human-animal chimeras. Debate on existing frameworks for the ethical assessment of chimeric animal research involving human tissue and their risk minimization has been documented[255,256]. In general, human-animal chimeras might be developed into a strategy to overcome organ shortage, but also a model for studies on organ development; pathogenesis, immunologic defenses, drug screening and toxicity testing. However, this requires us to overcome species-specific incompatibilities, such as differences in placental structures, cell cycle, and growth factor dependencies.
The production of meat in vitro using livestock PSCs is proposed as a clean and prominent alternative to slaughtering animals[257]. Bovine stem cells were used to make the world’s first burger from in vitro meat, which was served during a London press conference held in 2013[258]. This event was proclaimed as beneficial to reduce the global burden of the livestock industry, and was associated with environmental, ethical, and human health impacts[221,259]. The production of high-quality meat depends on the types of stem cells; source of ingredients and its composition. Among these, myoblast or satellite cells, and recently iPS cells are most important[260]. More recently, cattle umbilical cord blood cells have been reprogrammed to generated iPS cells, subsequently differentiated into muscle and fat cells[214,261]. Recent advancements have been made in the generation of stable bovine[145,262] and porcine PSCs[98], which could potentially be differentiated into skeletal muscle[263,264]. These PSCs form a cell bank with an unaltered and stable karyotype, and may eliminate further dependence on animals for cell isolation. However, this technology is in its infancy, and facing a number of challenges, such as whether in vitro meat will have the same taste as real meat, and whether this technology will be able to produce sufficient quantities in a cost-effective and clean way[265-267].
The PSCs comprise of ES cells derived from embryos and iPS cells obtained from reprogramming of somatic cells. The derivation of ES cells from embryos of livestock such as cattle, buffalo, sheep, goat, pig, horse, cat and dog had a relatively long and until recently unfruitful history. By contrast, the successful generation of iPS cells from livestock species is more promising and a straightforward technology. Commonly, the use of reprogramming vectors that integrate into the host cell genome, and are continuously expressed is a major bottleneck in the utility of iPS cells. To evade this hurdle, the use of non-integrating viral- and non-viral approaches for iPS cells generation resulted into safe and clinical grade cells for further downstream applications[82,268]. Various technical hurdles remain to be overcome for iPS cell technology to fully expand its potential, but remarkable achievements in recent years have led to clinical applications, provided new ways for the development of disease models, and improved patients’ treatments in a more adequate and personalized manner[269]. Looking ahead, the results of on-going clinical trials of iPS cells will deliver valuable information for preparing future strategies for cell-based therapy, drug testing, organ generation and disease modelling[237,270]. Pre-clinical testing of these approaches with livestock PSCs and large animal models are crucial for achieving these aims and successful translation into clinical therapies.
In future, PSCs along with novel upcoming technologies will synergically transform cellular reprogramming, differentiation and banking, which is necessarily connected to the industrialization of processes. More recently, massive progress has been achieved by the latest technologies in which ex-vivo and in vivo gene editing allowed efficient removal of the gene(s) responsible for the development of particular organs and the creation of new chimeric organs[271]. These approaches have the potential to innovate the field, but issues of safety and ethics need to be addressed in bringing the application of PSCs from bench to bedside.
Manuscript source: Invited manuscript
Specialty type: Cell Biology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Berezin AE, Wang H S-Editor: Fan JR L-Editor: Webster JR P-Editor: Xing YX
| 1. | Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19:662-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 2. | Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5956] [Cited by in F6Publishing: 5269] [Article Influence: 122.5] [Reference Citation Analysis (0)] |
| 3. | Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634-7638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3879] [Cited by in F6Publishing: 3463] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 4. | Nishikawa S, Jakt LM, Era T. Embryonic stem-cell culture as a tool for developmental cell biology. Nat Rev Mol Cell Biol. 2007;8:502-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 96] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 5. | Wu J, Platero-Luengo A, Sakurai M, Sugawara A, Gil MA, Yamauchi T, Suzuki K, Bogliotti YS, Cuello C, Morales Valencia M, Okumura D, Luo J, Vilariño M, Parrilla I, Soto DA, Martinez CA, Hishida T, Sánchez-Bautista S, Martinez-Martinez ML, Wang H, Nohalez A, Aizawa E, Martinez-Redondo P, Ocampo A, Reddy P, Roca J, Maga EA, Esteban CR, Berggren WT, Nuñez Delicado E, Lajara J, Guillen I, Guillen P, Campistol JM, Martinez EA, Ross PJ, Izpisua Belmonte JC. Interspecies Chimerism with Mammalian Pluripotent Stem Cells. Cell 2017; 168: 473-486. e15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 363] [Cited by in F6Publishing: 311] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 6. | Evans MJ, Notarianni E, Laurie S, Moor RM. Derivation and preliminary characterization of pluripotent cell lines from porcine and bovine blastocysts. Theriogenology. 1990, 33: 125-128. [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 105] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 7. | Piedrahita JA, Anderson GB, Bondurant RH. On the isolation of embryonic stem cells: Comparative behavior of murine, porcine and ovine embryos. Theriogenology. 1990;34:879-901. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 8. | Chen LR, Shiue YL, Bertolini L, Medrano JF, BonDurant RH, Anderson GB. Establishment of pluripotent cell lines from porcine preimplantation embryos. Theriogenology. 1999;52:195-212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 112] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Li M, Zhang D, Hou Y, Jiao L, Zheng X, Wang WH. Isolation and culture of embryonic stem cells from porcine blastocysts. Mol Reprod Dev. 2003;65:429-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | First NL, Sims MM, Park SP, Kent-First MJ. Systems for production of calves from cultured bovine embryonic cells. Reprod Fertil Dev. 1994;6:553-562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, Ponce de León FA, Robl JM. Transgenic bovine chimeric offspring produced from somatic cell-derived stem-like cells. Nat Biotechnol. 1998;16:642-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 241] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 12. | Mitalipova M, Beyhan Z, First NL. Pluripotency of bovine embryonic cell line derived from precompacting embryos. Cloning. 2001;3:59-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 88] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Yadav PS, Kues WA, Herrmann D, Carnwath JW, Niemann H. Bovine ICM derived cells express the Oct4 ortholog. Mol Reprod Dev. 2005;72:182-190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 14. | Notarianni E, Galli C, Laurie S, Moor RM, Evans MJ. Derivation of pluripotent, embryonic cell lines from the pig and sheep. J Reprod Fertil Suppl. 1991;43:255-260. [PubMed] [Cited in This Article: ] |
| 15. | Zhu SX, Sun Z, Zhang JP. Ovine (Ovis aries) blastula from an in vitro production system and isolation of primary embryonic stem cells. Zygote. 2007;15:35-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Behboodi E, Bondareva A, Begin I, Rao K, Neveu N, Pierson JT, Wylie C, Piero FD, Huang YJ, Zeng W, Tanco V, Baldassarre H, Karatzas CN, Dobrinski I. Establishment of goat embryonic stem cells from in vivo produced blastocyst-stage embryos. Mol Reprod Dev. 2011;78:202-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Kumar De A, Malakar D, Akshey YS, Jena MK, Dutta R. Isolation and characterization of embryonic stem cell-like cells from in vitro produced goat (Capra hircus) embryos. Anim Biotechnol. 2011;22:181-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Saito S, Ugai H, Sawai K, Yamamoto Y, Minamihashi A, Kurosaka K, Kobayashi Y, Murata T, Obata Y, Yokoyama K. Isolation of embryonic stem-like cells from equine blastocysts and their differentiation in vitro. FEBS Lett. 2002;531:389-396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Anand T, Kumar D, Singh MK, Shah RA, Chauhan MS, Manik RS, Singla SK, Palta P. Buffalo (Bubalus bubalis) embryonic stem cell-like cells and preimplantation embryos exhibit comparable expression of pluripotency-related antigens. Reprod Domest Anim. 2011;46:50-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Sharma R, George A, Kamble NM, Singh KP, Chauhan MS, Singla SK, Manik RS, Palta P. Optimization of culture conditions to support long-term self-renewal of buffalo (Bubalus bubalis) embryonic stem cell-like cells. Cell Reprogram. 2011;13:539-549. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Wang L, Duan E, Sung LY, Jeong BS, Yang X, Tian XC. Generation and characterization of pluripotent stem cells from cloned bovine embryos. Biol Reprod. 2005;73:149-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 99] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 22. | Kawase E, Yamazaki Y, Yagi T, Yanagimachi R, Pedersen RA. Mouse embryonic stem (ES) cell lines established from neuronal cell-derived cloned blastocysts. Genesis. 2000;28:156-163. [PubMed] [Cited in This Article: ] |
| 23. | Munsie MJ, Michalska AE, O'Brien CM, Trounson AO, Pera MF, Mountford PS. Isolation of pluripotent embryonic stem cells from reprogrammed adult mouse somatic cell nuclei. Curr Biol. 2000;10:989-992. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 277] [Cited by in F6Publishing: 282] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 24. | Wakayama S, Mizutani E, Kishigami S, Thuan NV, Ohta H, Hikichi T, Bui HT, Miyake M, Wakayama T. Mice cloned by nuclear transfer from somatic and ntES cells derived from the same individuals. J Reprod Dev. 2005;51:765-772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | George A, Sharma R, Singh KP, Panda SK, Singla SK, Palta P, Manik R, Chauhan MS. Production of cloned and transgenic embryos using buffalo (Bubalus bubalis) embryonic stem cell-like cells isolated from in vitro fertilized and cloned blastocysts. Cell Reprogram. 2011;13:263-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 440] [Cited by in F6Publishing: 346] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 27. | Wells DN, Oback B, Laible G. Cloning livestock: a return to embryonic cells. Trends Biotechnol. 2003;21:428-432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Kaufman MH, Robertson EJ, Handyside AH, Evans MJ. Establishment of pluripotential cell lines from haploid mouse embryos. J Embryol Exp Morphol. 1983;73:249-261. [PubMed] [Cited in This Article: ] |
| 29. | Wheeler MB. Development and validation of swine embryonic stem cells: a review. Reprod Fertil Dev. 1994;6:563-568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 126] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 30. | Strelchenko N. Bovine pluripotent stem cells. Theriogenology 1996; 45: 131–140 . [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 62] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Sritanaudomchai H, Pavasuthipaisit K, Kitiyanant Y, Kupradinun P, Mitalipov S, Kusamran T. Characterization and multilineage differentiation of embryonic stem cells derived from a buffalo parthenogenetic embryo. Mol Reprod Dev. 2007;74:1295-1302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Muzaffar M, Selokar NL, Singh KP, Zandi M, Singh MK, Shah RA, Chauhan MS, Singla SK, Palta P, Manik R. Equivalency of buffalo (Bubalus bubalis) embryonic stem cells derived from fertilized, parthenogenetic, and hand-made cloned embryos. Cell Reprogram. 2012;14:267-279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Singh KP, Kaushik R, Garg V, Sharma R, George A, Singh MK, Manik RS, Palta P, Singla SK, Chauhan MS. Expression pattern of pluripotent markers in different embryonic developmental stages of buffalo (Bubalus bubalis) embryos and putative embryonic stem cells generated by parthenogenetic activation. Cell Reprogram. 2012;14:530-538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 34. | Muñoz M, Rodríguez A, De Frutos C, Caamaño JN, Díez C, Facal N, Gómez E. Conventional pluripotency markers are unspecific for bovine embryonic-derived cell-lines. Theriogenology. 2008;69:1159-1164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 59] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 35. | Brevini TA, Pennarossa G, Gandolfi F. No shortcuts to pig embryonic stem cells. Theriogenology. 2010;74:544-550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 36. | Nowak-Imialek M, Kues W, Carnwath JW, Niemann H. Pluripotent stem cells and reprogrammed cells in farm animals. Microsc Microanal. 2011;17:474-497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 37. | Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, Pera MF, Ying QL. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299-1310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 522] [Cited by in F6Publishing: 499] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 38. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17989] [Cited by in F6Publishing: 17040] [Article Influence: 946.7] [Reference Citation Analysis (0)] |
| 39. | Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3332] [Cited by in F6Publishing: 3002] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 40. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14327] [Cited by in F6Publishing: 13571] [Article Influence: 848.2] [Reference Citation Analysis (0)] |
| 41. | Park ET, Gum JR, Kakar S, Kwon SW, Deng G, Kim YS. Aberrant expression of SOX2 upregulates MUC5AC gastric foveolar mucin in mucinous cancers of the colorectum and related lesions. Int J Cancer. 2008;122:1253-1260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 42. | Esteban MA, Xu J, Yang J, Peng M, Qin D, Li W, Jiang Z, Chen J, Deng K, Zhong M, Cai J, Lai L, Pei D. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2009;284:17634-17640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 320] [Cited by in F6Publishing: 328] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 43. | Ezashi T, Telugu BP, Alexenko AP, Sachdev S, Sinha S, Roberts RM. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci USA. 2009;106:10993-10998. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 362] [Cited by in F6Publishing: 336] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 44. | Wu Z, Chen J, Ren J, Bao L, Liao J, Cui C, Rao L, Li H, Gu Y, Dai H, Zhu H, Teng X, Cheng L, Xiao L. Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol. 2009;1:46-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 264] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 45. | West FD, Terlouw SL, Kwon DJ, Mumaw JL, Dhara SK, Hasneen K, Dobrinsky JR, Stice SL. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;19:1211-1220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 189] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 46. | Montserrat N, Bahima EG, Batlle L, Häfner S, Rodrigues AM, González F, Izpisúa Belmonte JC. Generation of pig iPS cells: a model for cell therapy. J Cardiovasc Transl Res. 2011;4:121-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 47. | Kues WA, Herrmann D, Barg-Kues B, Haridoss S, Nowak-Imialek M, Buchholz T, Streeck M, Grebe A, Grabundzija I, Merkert S, Martin U, Hall VJ, Rasmussen MA, Ivics Z, Hyttel P, Niemann H. Derivation and characterization of sleeping beauty transposon-mediated porcine induced pluripotent stem cells. Stem Cells Dev. 2013;22:124-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 48. | Bao L, He L, Chen J, Wu Z, Liao J, Rao L, Ren J, Li H, Zhu H, Qian L, Gu Y, Dai H, Xu X, Zhou J, Wang W, Cui C, Xiao L. Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 2011;21:600-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 49. | Li Y, Cang M, Lee AS, Zhang K, Liu D. Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PLoS One. 2011;6:e15947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 50. | Liu J, Balehosur D, Murray B, Kelly JM, Sumer H, Verma PJ. Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology 2012; 77: 338-46. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 51. | Ren J, Pak Y, He L, Qian L, Gu Y, Li H, Rao L, Liao J, Cui C, Xu X, Zhou J, Ri H, Xiao L. Generation of hircine-induced pluripotent stem cells by somatic cell reprogramming. Cell Res. 2011;21:849-853. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 52. | Song H, Li H, Huang M, Xu D, Gu C, Wang Z, Dong F, Wang F. Induced pluripotent stem cells from goat fibroblasts. Mol Reprod Dev. 2013;80:1009-1017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 53. | Shimada H, Nakada A, Hashimoto Y, Shigeno K, Shionoya Y, Nakamura T. Generation of canine induced pluripotent stem cells by retroviral transduction and chemical inhibitors. Mol Reprod Dev. 2010;77:2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 54. | Luo J, Suhr ST, Chang EA, Wang K, Ross PJ, Nelson LL, Venta PJ, Knott JG, Cibelli JB. Generation of leukemia inhibitory factor and basic fibroblast growth factor-dependent induced pluripotent stem cells from canine adult somatic cells. Stem Cells Dev. 2011;20:1669-1678. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 55. | Whitworth DJ, Ovchinnikov DA, Wolvetang EJ. Generation and characterization of LIF-dependent canine induced pluripotent stem cells from adult dermal fibroblasts. Stem Cells Dev. 2012;21:2288-2297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 56. | Koh S, Thomas R, Tsai S, Bischoff S, Lim JH, Breen M, Olby NJ, Piedrahita JA. Growth requirements and chromosomal instability of induced pluripotent stem cells generated from adult canine fibroblasts. Stem Cells Dev. 2013;22:951-963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 57. | Sumer H, Liu J, Malaver-Ortega LF, Lim ML, Khodadadi K, Verma PJ. NANOG is a key factor for induction of pluripotency in bovine adult fibroblasts. J Anim Sci. 2011;89:2708-2716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 58. | Talluri TR, Kumar D, Glage S, Garrels W, Ivics Z, Debowski K, Behr R, Niemann H, Kues WA. Derivation and characterization of bovine induced pluripotent stem cells by transposon-mediated reprogramming. Cell Reprogram. 2015;17:131-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 59. | Nagy K, Sung HK, Zhang P, Laflamme S, Vincent P, Agha-Mohammadi S, Woltjen K, Monetti C, Michael IP, Smith LC, Nagy A. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev Rep. 2011;7:693-702. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 165] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 60. | Hackett CH, Greve L, Novakofski KD, Fortier LA. Comparison of gene-specific DNA methylation patterns in equine induced pluripotent stem cell lines with cells derived from equine adult and fetal tissues. Stem Cells Dev. 2012;21:1803-1811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 61. | Breton A, Sharma R, Diaz AC, Parham AG, Graham A, Neil C, Whitelaw CB, Milne E, Donadeu FX. Derivation and characterization of induced pluripotent stem cells from equine fibroblasts. Stem Cells Dev. 2013;22:611-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 62. | Donadeu FX. Equine induced pluripotent stem cells or how to turn skin cells into neurons: horse tissues a la carte? Equine Vet J. 2014;46:534-537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 63. | Whitworth DJ, Ovchinnikov DA, Sun J, Fortuna PR, Wolvetang EJ. Generation and characterization of leukemia inhibitory factor-dependent equine induced pluripotent stem cells from adult dermal fibroblasts. Stem Cells Dev. 2014;23:1515-1523. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 64. | Sharma R, Livesey MR, Wyllie DJ, Proudfoot C, Whitelaw CB, Hay DC, Donadeu FX. Generation of functional neurons from feeder-free, keratinocyte-derived equine induced pluripotent stem cells. Stem Cells Dev. 2014;23:1524-1534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 65. | Deng Y, Liu Q, Luo C, Chen S, Li X, Wang C, Liu Z, Lei X, Zhang H, Sun H, Lu F, Jiang J, Shi D. Generation of induced pluripotent stem cells from buffalo (Bubalus bubalis) fetal fibroblasts with buffalo defined factors. Stem Cells Dev. 2012;21:2485-2494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 66. | Kumar D, Anand T, Vijayalakshmy K, Sharma P, Rajendran R, Selokar NL, Yadav PS, Kumar D. Transposon mediated reprogramming of buffalo fetal fibroblasts to induced pluripotent stem cells in feeder free culture conditions. Res Vet Sci. 2019;123:252-260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 67. | Sartori C, DiDomenico AI, Thomson AJ, Milne E, Lillico SG, Burdon TG, Whitelaw CB. Ovine-induced pluripotent stem cells can contribute to chimeric lambs. Cell Reprogram. 2012;14:8-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 68. | Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, Toyoshima M, Niwa O, Oshimura M, Heike T, Nakahata T, Ishino F, Ogura A, Shinohara T. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001-1012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 618] [Cited by in F6Publishing: 561] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 69. | Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, Hasenfuss G. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199-1203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 650] [Cited by in F6Publishing: 596] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 70. | Lee SW, Wu G, Choi NY, Lee HJ, Bang JS, Lee Y, Lee M, Ko K, Schöler HR, Ko K. Self-Reprogramming of Spermatogonial Stem Cells into Pluripotent Stem Cells without Microenvironment of Feeder Cells. Mol Cells. 2018;41:631-638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 6] [Reference Citation Analysis (0)] |
| 71. | Ko K, Tapia N, Wu G, Kim JB, Bravo MJ, Sasse P, Glaser T, Ruau D, Han DW, Greber B, Hausdörfer K, Sebastiano V, Stehling M, Fleischmann BK, Brüstle O, Zenke M, Schöler HR. Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell. 2009;5:87-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 205] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 72. | Wang H, Jiang M, Bi H, Chen X, He L, Li X, Wu J. Conversion of female germline stem cells from neonatal and prepubertal mice into pluripotent stem cells. J Mol Cell Biol. 2014;6:164-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 73. | Zeng F, Huang F, Guo J, Hu X, Liu C, Wang H. Emerging methods to generate artificial germ cells from stem cells. Biol Reprod. 2015;92:89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 74. | Ben-Nun IF, Montague SC, Houck ML, Tran HT, Garitaonandia I, Leonardo TR, Wang YC, Charter SJ, Laurent LC, Ryder OA, Loring JF. Induced pluripotent stem cells from highly endangered species. Nat Methods. 2011;8:829-831. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 115] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 75. | Schmidt R, Plath K. The roles of the reprogramming factors Oct4, Sox2 and Klf4 in resetting the somatic cell epigenome during induced pluripotent stem cell generation. Genome Biol. 2012;13:251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 76. | Rashid T, Kobayashi T, Nakauchi H. Revisiting the flight of Icarus: making human organs from PSCs with large animal chimeras. Cell Stem Cell. 2014;15:406-409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 77. | Keefer CL. Artificial cloning of domestic animals. Proc Natl Acad Sci U S A. 2015;112:8874-8878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 78. | Kumar D, Talluri TR, Anand T, Kues WA. Induced pluripotent stem cells: Mechanisms, achievements and perspectives in farm animals. World J Stem Cells. 2015;7:315-328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 79. | Ogorevc J, Orehek S, Dovč P. Cellular reprogramming in farm animals: an overview of iPSC generation in the mammalian farm animal species. J Anim Sci Biotechnol. 2016;7:10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 80. | Kumar D, Anand T, Kues WA. Clinical potential of human-induced pluripotent stem cells : Perspectives of induced pluripotent stem cells. Cell Biol Toxicol. 2017;33:99-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 81. | Kumar D, Talluri TR, Anand T, Kues WA. Transposon-based reprogramming to induced pluripotency. Histol Histopathol. 2015;30:1397-1409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 82. | Haridhasapavalan KK, Borgohain MP, Dey C, Saha B, Narayan G, Kumar S, Thummer RP. An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene. 2019;686:146-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 83. | Kumar D, Anand T, Talluri TR, Kues WA. Potential of transposon-mediated cellular reprogramming towards cell-based therapies. World J Stem Cells. 2020;12:527-544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 84. | Koh S, Piedrahita JA. From "ES-like" cells to induced pluripotent stem cells: a historical perspective in domestic animals. Theriogenology. 2014;81:103-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 85. | Ezashi T, Yuan Y, Roberts RM. Pluripotent Stem Cells from Domesticated Mammals. Annu Rev Anim Biosci. 2016;4:223-253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 86. | Coucouvanis E, Martin GR. BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development. 1999;126:535-546. [PubMed] [Cited in This Article: ] |
| 87. | Muñoz M, Díez C, Caamaño JN, Jouneau A, Hue I, Gómez E. Embryonic stem cells in cattle. Reprod Domest Anim. 2008;43 Suppl 4:32-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 88. | Talbot NC, Blomberg Le Ann. The pursuit of ES cell lines of domesticated ungulates. Stem Cell Rev. 2008;4:235-254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 89. | Gandolfi F, Pennarossa G, Maffei S, Brevini T. Why is it so difficult to derive pluripotent stem cells in domestic ungulates? Reprod Domest Anim. 2012;47 Suppl 5:11-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 90. | Maddox-Hyttel P, Alexopoulos NI, Vajta G, Lewis I, Rogers P, Cann L, Callesen H, Tveden-Nyborg P, Trounson A. Immunohistochemical and ultrastructural characterization of the initial post-hatching development of bovine embryos. Reproduction. 2003;125:607-623. [PubMed] [Cited in This Article: ] |
| 91. | Vejlsted M, Du Y, Vajta G, Maddox-Hyttel P. Post-hatching development of the porcine and bovine embryo--defining criteria for expected development in vivo and in vitro. Theriogenology. 2006;65:153-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 70] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 92. | Blomberg LA, Telugu BP. Twenty years of embryonic stem cell research in farm animals. Reprod Domest Anim. 2012;47 Suppl 4:80-85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 93. | Degrelle SA, Campion E, Cabau C, Piumi F, Reinaud P, Richard C, Renard JP, Hue I. Molecular evidence for a critical period in mural trophoblast development in bovine blastocysts. Dev Biol. 2005;288:448-460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 159] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 94. | Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod Fertil Dev. 2007;19:111-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 92] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 95. | Gonçalves NN, Ambrósio CE, Piedrahita JA. Stem cells and regenerative medicine in domestic and companion animals: a multispecies perspective. Reprod Domest Anim. 2014;49 Suppl 4:2-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 96. | Vassiliev I, Vassilieva S, Beebe LF, Harrison SJ, McIlfatrick SM, Nottle MB. In vitro and in vivo characterization of putative porcine embryonic stem cells. Cell Reprogram. 2010;12:223-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 97. | Xue B, Li Y, He Y, Wei R, Sun R, Yin Z, Bou G, Liu Z. Porcine Pluripotent Stem Cells Derived from IVF Embryos Contribute to Chimeric Development In Vivo. PLoS One. 2016;11:e0151737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 98. | Gao X, Nowak-Imialek M, Chen X, Chen D, Herrmann D, Ruan D, Chen ACH, Eckersley-Maslin MA, Ahmad S, Lee YL, Kobayashi T, Ryan D, Zhong J, Zhu J, Wu J, Lan G, Petkov S, Yang J, Antunes L, Campos LS, Fu B, Wang S, Yong Y, Wang X, Xue SG, Ge L, Liu Z, Huang Y, Nie T, Li P, Wu D, Pei D, Zhang Y, Lu L, Yang F, Kimber SJ, Reik W, Zou X, Shang Z, Lai L, Surani A, Tam PPL, Ahmed A, Yeung WSB, Teichmann SA, Niemann H, Liu P. Establishment of porcine and human expanded potential stem cells. Nat Cell Biol. 2019;21:687-699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 217] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 99. | Vaags AK, Rosic-Kablar S, Gartley CJ, Zheng YZ, Chesney A, Villagómez DA, Kruth SA, Hough MR. Derivation and characterization of canine embryonic stem cell lines with in vitro and in vivo differentiation potential. Stem Cells. 2009;27:329-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 100. | Wang H, Pei Y, Li N, Han J. Progress, problems and prospects of porcine pluripotent stem cells. Front Agric Sci Eng. 2014;1:6-15. [DOI] [Cited in This Article: ] |
| 101. | Malaver-Ortega LF, Sumer H, Liu J, Verma PJ. The state of the art for pluripotent stem cells derivation in domestic ungulates. Theriogenology. 2012;78:1749-1762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 102. | Nowak-Imialek M, Niemann H. Pluripotent cells in farm animals: state of the art and future perspectives. Reprod Fertil Dev. 2012;25:103-128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 103. | Navarro M, Soto DA, Pinzon CA, Wu J, Ross PJ. Livestock pluripotency is finally captured in vitro. Reprod Fertil Dev. 2019;32:11-39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 104. | Lu Y, Mumaw JL, West FD, Stice SL. Livestock induced pluripotent stem cells. Reprod Domest Anim. 2012;47 Suppl 4:72-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 105. | West FD, Uhl EW, Liu Y, Stowe H, Lu Y, Yu P, Gallegos-Cardenas A, Pratt SL, Stice SL. Brief report: chimeric pigs produced from induced pluripotent stem cells demonstrate germline transmission and no evidence of tumor formation in young pigs. Stem Cells. 2011;29:1640-1643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 106. | Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567-582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1088] [Cited by in F6Publishing: 988] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 107. | Cai J, Li W, Su H, Qin D, Yang J, Zhu F, Xu J, He W, Guo X, Labuda K, Peterbauer A, Wolbank S, Zhong M, Li Z, Wu W, So KF, Redl H, Zeng L, Esteban MA, Pei D. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J Biol Chem. 2010;285:11227-11234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 108. | Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379-391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2554] [Cited by in F6Publishing: 2441] [Article Influence: 93.9] [Reference Citation Analysis (0)] |
| 109. | Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1695] [Cited by in F6Publishing: 1634] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 110. | Hart AH, Hartley L, Ibrahim M, Robb L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev Dyn. 2004;230:187-198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 229] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 111. | Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049-1061. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1052] [Cited by in F6Publishing: 1039] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 112. | Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, Mongan NP. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093-1108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 327] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 113. | Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031-6046. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 502] [Cited by in F6Publishing: 505] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 114. | Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947-956. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3309] [Cited by in F6Publishing: 3271] [Article Influence: 172.2] [Reference Citation Analysis (0)] |
| 115. | Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997-1001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 277] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 116. | Hayashi Y, Caboni L, Das D, Yumoto F, Clayton T, Deller MC, Nguyen P, Farr CL, Chiu HJ, Miller MD, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Tomoda K, Conklin BR, Wilson IA, Yamanaka S, Fletterick RJ. Structure-based discovery of NANOG variant with enhanced properties to promote self-renewal and reprogramming of pluripotent stem cells. Proc Natl Acad Sci U S A. 2015;112:4666-4671. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 117. | Kues WA, Nowak-Imialek M, Haridoss S, Niemann H. Strategies for the derivation of pluripotent cells from farm animals. Reprod Domest Anim. 2010;45 Suppl 3:25-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 118. | Han X, Han J, Ding F, Cao S, Lim SS, Dai Y, Zhang R, Zhang Y, Lim B, Li N. Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 2011;21:1509-1512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 119. | Cao H, Yang P, Pu Y, Sun X, Yin H, Zhang Y, Zhang Y, Li Y, Liu Y, Fang F, Zhang Z, Tao Y, Zhang X. Characterization of bovine induced pluripotent stem cells by lentiviral transduction of reprogramming factor fusion proteins. Int J Biol Sci. 2012;8:498-511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 120. | Canizo JR, Vazquez Echegaray C, Klisch D, Aller JF, Paz DA, Alberio RH, Alberio R, Guberman AS. Exogenous human OKSM factors maintain pluripotency gene expression of bovine and porcine iPS-like cells obtained with STEMCCA delivery system. BMC Res Notes. 2018;11:509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 121. | German SD, Campbell KH, Thornton E, McLachlan G, Sweetman D, Alberio R. Ovine induced pluripotent stem cells are resistant to reprogramming after nuclear transfer. Cell Reprogram. 2015;17:19-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 122. | Dutton LC, Dudhia J, Guest DJ, Connolly DJ. Inducing Pluripotency in the Domestic Cat (Felis catus). Stem Cells Dev. 2019;28:1299-1309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 123. | Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212-215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1027] [Cited by in F6Publishing: 961] [Article Influence: 73.9] [Reference Citation Analysis (0)] |
| 124. | Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1024] [Cited by in F6Publishing: 925] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 125. | Tashiro K, Inamura M, Kawabata K, Sakurai F, Yamanishi K, Hayakawa T, Mizuguchi H. Efficient adipocyte and osteoblast differentiation from mouse induced pluripotent stem cells by adenoviral transduction. Stem Cells. 2009;27:1802-1811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 126. | Chow L, Johnson V, Regan D, Wheat W, Webb S, Koch P, Dow S. Safety and immune regulatory properties of canine induced pluripotent stem cell-derived mesenchymal stem cells. Stem Cell Res. 2017;25:221-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 127. | Tsukamoto M, Nishimura T, Yodoe K, Kanegi R, Tsujimoto Y, Alam ME, Kuramochi M, Kuwamura M, Ohtaka M, Nishimura K, Nakanishi M, Inaba T, Sugiura K, Hatoya S. Generation of Footprint-Free Canine Induced Pluripotent Stem Cells Using Auto-Erasable Sendai Virus Vector. Stem Cells Dev. 2018;27:1577-1586. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 128. | Huang B, Li T, Alonso-Gonzalez L, Gorre R, Keatley S, Green A, Turner P, Kallingappa PK, Verma V, Oback B. A virus-free poly-promoter vector induces pluripotency in quiescent bovine cells under chemically defined conditions of dual kinase inhibition. PLoS One. 2011;6:e24501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 129. | Kawaguchi T, Tsukiyama T, Kimura K, Matsuyama S, Minami N, Yamada M, Imai H. Generation of Naïve Bovine Induced Pluripotent Stem Cells Using PiggyBac Transposition of Doxycycline-Inducible Transcription Factors. PLoS One. 2015;10:e0135403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 130. | Zhao L, Wang Z, Zhang J, Yang J, Gao X, Wu B, Zhao G, Bao S, Hu S, Liu P, Li X. Characterization of the single-cell derived bovine induced pluripotent stem cells. Tissue Cell. 2017;49:521-527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 131. | Telugu BP, Ezashi T, Roberts RM. Porcine induced pluripotent stem cells analogous to naïve and primed embryonic stem cells of the mouse. Int J Dev Biol. 2010;54:1703-1711. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 132. | Li D, Secher J, Hyttel P, Ivask M, Kolko M, Hall VJ, Freude KK. Generation of transgene-free porcine intermediate type induced pluripotent stem cells. Cell Cycle. 2018;17:2547-2563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 133. | Pessôa LVF, Bressan FF, Freude KK. Induced pluripotent stem cells throughout the animal kingdom: Availability and applications. World J Stem Cells. 2019;11:491-505. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 134. | Goszczynski DE, Cheng H, Demyda-Peyrás S, Medrano JF, Wu J, Ross PJ. In vitro breeding: application of embryonic stem cells to animal production†. Biol Reprod. 2019;100:885-895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 135. | Ying Y, Liu XM, Marble A, Lawson KA, Zhao GQ. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol. 2000;14:1053-1063. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 267] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 136. | Ying Y, Zhao GQ. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev Biol. 2001;232:484-492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 300] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 137. | Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571-584. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 381] [Cited by in F6Publishing: 361] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 138. | García-Ruiz A, Cole JB, VanRaden PM, Wiggans GR, Ruiz-López FJ, Van Tassell CP. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc Natl Acad Sci USA. 2016;113:E3995-E4004. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 295] [Cited by in F6Publishing: 268] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 139. | Hou Z, An L, Han J, Yuan Y, Chen D, Tian J. Revolutionize livestock breeding in the future: an animal embryo-stem cell breeding system in a dish. J Anim Sci Biotechnol. 2018;9:90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 140. | Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 930] [Cited by in F6Publishing: 938] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 141. | Hayashi K, Ogushi S, Kurimoto K, Shimamoto S, Ohta H, Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971-975. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 510] [Cited by in F6Publishing: 495] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 142. | Teramura T, Takehara T, Kawata N, Fujinami N, Mitani T, Takenoshita M, Matsumoto K, Saeki K, Iritani A, Sagawa N, Hosoi Y. Primate embryonic stem cells proceed to early gametogenesis in vitro. Cloning Stem Cells. 2007;9:144-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 143. | Yamauchi K, Hasegawa K, Chuma S, Nakatsuji N, Suemori H. In vitro germ cell differentiation from cynomolgus monkey embryonic stem cells. PLoS One. 2009;4:e5338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 144. | Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, Valli H, Rodriguez M, Ezzelarab M, Dargo G, Peterson K, Masterson K, Ramsey C, Ward T, Lienesch M, Volk A, Cooper DK, Thomson AW, Kiss JE, Penedo MC, Schatten GP, Mitalipov S, Orwig KE. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715-726. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 319] [Cited by in F6Publishing: 272] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 145. | Bogliotti YS, Wu J, Vilarino M, Okamura D, Soto DA, Zhong C, Sakurai M, Sampaio RV, Suzuki K, Izpisua Belmonte JC, Ross PJ. Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc Natl Acad Sci USA. 2018;115:2090-2095. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 146. | Li Y, Wang X, Feng X, Liao S, Zhang D, Cui X, Gao F, Han C. Generation of male germ cells from mouse induced pluripotent stem cells in vitro. Stem Cell Res. 2014;12:517-530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 147. | Wang H, Xiang J, Zhang W, Li J, Wei Q, Zhong L, Ouyang H, Han J. Induction of Germ Cell-like Cells from Porcine Induced Pluripotent Stem Cells. Sci Rep. 2016;6:27256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 148. | Kobayashi T, Zhang H, Tang WWC, Irie N, Withey S, Klisch D, Sybirna A, Dietmann S, Contreras DA, Webb R, Allegrucci C, Alberio R, Surani MA. Principles of early human development and germ cell program from conserved model systems. Nature. 2017;546:416-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 149. | Malaver-Ortega LF, Sumer H, Jain K, Verma PJ. Bone morphogenetic protein 4 and retinoic acid trigger bovine VASA homolog expression in differentiating bovine induced pluripotent stem cells. Mol Reprod Dev. 2016;83:149-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 150. | Xie B, Qin Z, Huang B, Xie T, Yao H, Wei Y, Yang X, Shi D, Jiang H. In vitro culture and differentiation of buffalo (Bubalus bubalis) spermatogonia. Reprod Domest Anim. 2010;45:275-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 151. | Shah SM, Singla SK, Palta P, Manik RS, Chauhan MS. Retinoic acid induces differentiation of buffalo (Bubalus bubalis) embryonic stem cells into germ cells. Gene. 2017;626:358-366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 152. | Zhu Y, Hu HL, Li P, Yang S, Zhang W, Ding H, Tian RH, Ning Y, Zhang LL, Guo XZ, Shi ZP, Li Z, He Z. Generation of male germ cells from induced pluripotent stem cells (iPS cells): an in vitro and in vivo study. Asian J Androl. 2012;14:574-579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 153. | Cui T, Li Z, Zhou Q, Li W. Current advances in haploid stem cells. Protein Cell. 2020;11:23-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 154. | Li Y, Shuai L. A versatile genetic tool: haploid cells. Stem Cell Res Ther. 2017;8:197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 155. | Bai M, Wu Y, Li J. Generation and application of mammalian haploid embryonic stem cells. J Intern Med. 2016;280:236-245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 156. | Soto DA, Ross PJ. Pluripotent stem cells and livestock genetic engineering. Transgenic Res. 2016;25:289-306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 157. | Taft RA, Low BE, Byers SL, Murray SA, Kutny P, Wiles MV. The perfect host: a mouse host embryo facilitating more efficient germ line transmission of genetically modified embryonic stem cells. PLoS One. 2013;8:e67826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 158. | Laible G, Alonso-González L. Gene targeting from laboratory to livestock: current status and emerging concepts. Biotechnol J. 2009;4:1278-1292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 159. | Ohtsuka M, Miura H, Nakaoka H, Kimura M, Sato M, Inoko H. Targeted transgenesis through pronuclear injection of improved vectors into in vitro fertilized eggs. Transgenic Res. 2012;21:225-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 160. | Tang L, González R, Dobrinski I. Germline modification of domestic animals. Anim Reprod. 2015;12:93-104. [PubMed] [Cited in This Article: ] |
| 161. | Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, Vargas F, Sathiyaseelan J, Wu H, Matsushita H, Koster J, Kato S, Ishida I, Soto C, Robl JM, Kuroiwa Y. Production of cattle lacking prion protein. Nat Biotechnol. 2007;25:132-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 204] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 162. | Fontes A, Lakshmipathy U. Advances in genetic modification of pluripotent stem cells. Biotechnol Adv. 2013;31:994-1001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 163. | Kou Z, Kang L, Yuan Y, Tao Y, Zhang Y, Wu T, He J, Wang J, Liu Z, Gao S. Mice cloned from induced pluripotent stem cells (iPSCs). Biol Reprod. 2010;83:238-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 164. | Fan N, Chen J, Shang Z, Dou H, Ji G, Zou Q, Wu L, He L, Wang F, Liu K, Liu N, Han J, Zhou Q, Pan D, Yang D, Zhao B, Ouyang Z, Liu Z, Zhao Y, Lin L, Zhong C, Wang Q, Wang S, Xu Y, Luan J, Liang Y, Yang Z, Li J, Lu C, Vajta G, Li Z, Ouyang H, Wang H, Wang Y, Yang Y, Liu Z, Wei H, Luan Z, Esteban MA, Deng H, Yang H, Pei D, Li N, Pei G, Liu L, Du Y, Xiao L, Lai L. Piglets cloned from induced pluripotent stem cells. Cell Res. 2013;23:162-166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 165. | Xie B, Wang J, Liu S, Wang J, Xue B, Li J, Wei R, Zhao Y, Liu Z. Positive correlation between the efficiency of induced pluripotent stem cells and the development rate of nuclear transfer embryos when the same porcine embryonic fibroblast lines are used as donor cells. Cell Reprogram. 2014;16:206-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 166. | Zhou S, Ding C, Zhao X, Wang E, Dai X, Liu L, Li W, Liu Z, Wan H, Feng C, Hai T, Wang L, Zhou Q. Successful generation of cloned mice using nuclear transfer from induced pluripotent stem cells. Cell Res. 2010;20:850-853. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 167. | Petersen B. Basics of genome editing technology and its application in livestock species. Reprod Domest Anim. 2017;52 Suppl 3:4-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 168. | Wu G, Bazer FW. Application of new biotechnologies for improvements in swine nutrition and pork production. J Anim Sci Biotechnol. 2019;10:28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 169. | Zhao J, Lai L, Ji W, Zhou Q. Genome editing in large animals: current status and future prospects. Natl Sci Rev. 2019;6: 402-420. [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 170. | Kumar D, Kues WA Application of genome editing in farm animals. In: Genomics and biotechnological advances in veterinary, poultry, and fisheries. Academic Press. 2019;131-149. [DOI] [Cited in This Article: ] |
| 171. | Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5:1. [DOI] [Cited in This Article: ] [Cited by in Crossref: 949] [Cited by in F6Publishing: 839] [Article Influence: 209.8] [Reference Citation Analysis (0)] |
| 172. | Park KE, Telugu BP. Role of stem cells in large animal genetic engineering in the TALENs-CRISPR era. Reprod Fertil Dev. 2013;26:65-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 173. | Gribben JG. Stem cell transplantation in chronic lymphocytic leukemia. Biol Blood Marrow Transplant. 2009;15:53-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 174. | Harding J, Roberts RM, Mirochnitchenko O. Large animal models for stem cell therapy. Stem Cell Res Ther. 2013;4:23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 113] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 175. | Volk SW, Theoret C. Translating stem cell therapies: the role of companion animals in regenerative medicine. Wound Repair Regen. 2013;21:382-394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 176. | Dehghan MM, Baghaban Eslaminejad M, Motallebizadeh N, Ashrafi Halan J, Tagiyar L, Soroori S, Nikmahzar A, Pedram M, Shahverdi A, Kazemi Mehrjerdi H, Izadi S. Transplantation of Autologous Bone Marrow Mesenchymal Stem Cells with Platelet-Rich Plasma Accelerate Distraction Osteogenesis in A Canine Model. Cell J. 2015;17:243-252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 14] [Reference Citation Analysis (0)] |
| 177. | Kastner A, Gauthier P. Are rodents an appropriate pre-clinical model for treating spinal cord injury? Exp Neurol. 2008;213:249-256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 178. | Kehinde EO. They see a rat, we seek a cure for diseases: the current status of animal experimentation in medical practice. Med Princ Pract. 2013;22 Suppl 1:52-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 179. | Gün G, Kues WA. Current progress of genetically engineered pig models for biomedical research. Biores Open Access. 2014;3:255-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 180. | Cong X, Zhang SM, Ellis MW, Luo J. Large Animal Models for the Clinical Application of Human Induced Pluripotent Stem Cells. Stem Cells Dev. 2019;28:1288-1298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 181. | Plews JR, Gu M, Longaker MT, Wu JC. Large animal induced pluripotent stem cells as pre-clinical models for studying human disease. J Cell Mol Med. 2012;16:1196-1202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 182. | Kurome M, Geistlinger L, Kessler B, Zakhartchenko V, Klymiuk N, Wuensch A, Richter A, Baehr A, Kraehe K, Burkhardt K, Flisikowski K, Flisikowska T, Merkl C, Landmann M, Durkovic M, Tschukes A, Kraner S, Schindelhauer D, Petri T, Kind A, Nagashima H, Schnieke A, Zimmer R, Wolf E. Factors influencing the efficiency of generating genetically engineered pigs by nuclear transfer: multi-factorial analysis of a large data set. BMC Biotechnol. 2013;13:43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 183. | Bassols A, Costa C, Eckersall PD, Osada J, Sabrià J, Tibau J. The pig as an animal model for human pathologies: A proteomics perspective. Proteomics Clin Appl. 2014;8:715-731. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 158] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 184. | Madeja ZE, Pawlak P, Piliszek A. Beyond the mouse: non-rodent animal models for study of early mammalian development and biomedical research. Int J Dev Biol. 2019;63:187-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 185. | Duranthon V, Beaujean N, Brunner M, Odening KE, Santos AN, Kacskovics I, Hiripi L, Weinstein EJ, Bosze Z. On the emerging role of rabbit as human disease model and the instrumental role of novel transgenic tools. Transgenic Res. 2012;21:699-713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 186. | Cebrian-Serrano A, Stout T, Dinnyes A. Veterinary applications of induced pluripotent stem cells: regenerative medicine and models for disease? Vet J. 2013;198:34-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 187. | Paterson YZ, Kafarnik C, Guest DJ. Characterization of companion animal pluripotent stem cells. Cytometry A. 2018;93:137-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 188. | Kues WA, Niemann H. The contribution of farm animals to human health. Trends Biotechnol. 2004;22:286-294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 189. | Klymiuk N, Seeliger F, Bohlooly-Y M, Blutke A, Rudmann DG, Wolf E. Tailored Pig Models for Preclinical Efficacy and Safety Testing of Targeted Therapies. Toxicol Pathol. 2016;44:346-357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 190. | Zhou L, Wang W, Liu Y, Fernandez de Castro J, Ezashi T, Telugu BP, Roberts RM, Kaplan HJ, Dean DC. Differentiation of induced pluripotent stem cells of swine into rod photoreceptors and their integration into the retina. Stem Cells. 2011;29:972-980. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 191. | Amado LC, Schuleri KH, Saliaris AP, Boyle AJ, Helm R, Oskouei B, Centola M, Eneboe V, Young R, Lima JA, Lardo AC, Heldman AW, Hare JM. Multimodality noninvasive imaging demonstrates in vivo cardiac regeneration after mesenchymal stem cell therapy. J Am Coll Cardiol. 2006;48:2116-2124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 139] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 192. | Gandolfi F, Vanelli A, Pennarossa G, Rahaman M, Acocella F, Brevini TA. Large animal models for cardiac stem cell therapies. Theriogenology. 2011;75:1416-1425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 193. | Mazhari R, Hare JM. Translational findings from cardiovascular stem cell research. Trends Cardiovasc Med. 2012;22:1-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 194. | Ye L, Chang YH, Xiong Q, Zhang P, Zhang L, Somasundaram P, Lepley M, Swingen C, Su L, Wendel JS, Guo J, Jang A, Rosenbush D, Greder L, Dutton JR, Zhang J, Kamp TJ, Kaufman DS, Ge Y, Zhang J. Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell Stem Cell. 2014;15:750-761. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 337] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 195. | Gu M, Nguyen PK, Lee AS, Xu D, Hu S, Plews JR, Han L, Huber BC, Lee WH, Gong Y, de Almeida PE, Lyons J, Ikeno F, Pacharinsak C, Connolly AJ, Gambhir SS, Robbins RC, Longaker MT, Wu JC. Microfluidic single-cell analysis shows that porcine induced pluripotent stem cell-derived endothelial cells improve myocardial function by paracrine activation. Circ Res. 2012;111:882-893. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 196. | van der Spoel TI, Jansen of Lorkeers SJ, Agostoni P, van Belle E, Gyöngyösi M, Sluijter JP, Cramer MJ, Doevendans PA, Chamuleau SA. Human relevance of pre-clinical studies in stem cell therapy: systematic review and meta-analysis of large animal models of ischaemic heart disease. Cardiovasc Res. 2011;91:649-658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 185] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 197. | Guo J, Wang H, Hu X. Reprogramming and transdifferentiation shift the landscape of regenerative medicine. DNA Cell Biol. 2013;32:565-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 198. | Holt WV, Brown JL, Comizzoli P. Reproductive science as an essential component of conservation biology. Adv Exp Med Biol. 2014;753:3-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 199. | Brose U, Hillebrand H. Biodiversity and ecosystem functioning in dynamic landscapes. Philos Trans R Soc Lond B Biol Sci. 2016;371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 200. | Comizzoli P, Mermillod P, Mauget R. Reproductive biotechnologies for endangered mammalian species. Reprod Nutr Dev. 2000;40:493-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 82] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 201. | Mara L, Casu S, Carta A, Dattena M. Cryobanking of farm animal gametes and embryos as a means of conserving livestock genetics. Anim Reprod Sci. 2013;138:25-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 202. | Comizzoli P, Holt WV. Recent advances and prospects in germplasm preservation of rare and endangered species. Adv Exp Med Biol. 2014;753:331-356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 203. | Arat S, Caputcu AT, Akkoc T, Pabuccuoglu S, Sagirkaya H, Cirit U, Nak Y, Koban E, Bagis H, Demir K, Nak D, Senunver A, Kilicaslan R, Tuna B, Cetinkaya G, Denizci M, Aslan O. Using cell banks as a tool in conservation programmes of native domestic breeds: the production of the first cloned Anatolian Grey cattle. Reprod Fertil Dev. 2011;23:1012-1023. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 204. | Groeneveld LF, Gregusson S, Guldbrandtsen B, Hiemstra SJ, Hveem K, Kantanen J, Lohi H, Stroemstedt L, Berg P. Domesticated Animal Biobanking: Land of Opportunity. PLoS Biol. 2016;14:e1002523. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 205. | Selokar NL, Sharma P, Krishna A, Kumar D, Kumar D, Saini M, Sharma A, Vijayalakshmy K, Yadav PS. Establishment of a Somatic Cell Bank for Indian Buffalo Breeds and Assessing the Suitability of the Cryopreserved Cells for Somatic Cell Nuclear Transfer. Cell Reprogram. 2018;20:157-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 206. | Mastromonaco GF, González-Grajales LA, Filice M, Comizzoli P. Somatic cells, stem cells, and induced pluripotent stem cells: how do they now contribute to conservation? Adv Exp Med Biol. 2014;753:385-427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 207. | Comizzoli P, Holt WV. Implications of the Nagoya Protocol for genome resource banks composed of biomaterials from rare and endangered species. Reprod Fertil Dev. 2016;28:1145-1160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 208. | Comizzoli P, Holt WV. Breakthroughs and new horizons in reproductive biology of rare and endangered animal species. Biol Reprod. 2019;101:514-525. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 209. | Oh HY, Jin X, Kim JG, Oh MJ, Pian X, Kim JM, Yoon MS, Son CI, Lee YS, Hong KC, Kim H, Choi YJ, Whang KY. Characteristics of primary and immortalized fibroblast cells derived from the miniature and domestic pigs. BMC Cell Biol. 2007;8:20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 210. | Liu C, Guo Y, Guan W, Ma Y, Zhang HH, Tang X. Establishment and biological characteristics of Luxi cattle fibroblast bank. Tissue Cell. 2008;40:417-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 211. | Liu B, Zhang H, Hao M, Yu S. Establishment and characterization of two fetal fibroblast cell lines from the yak. In Vitro Cell Dev Biol Anim. 2012;48:619-624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 212. | Singh M, Sharma AK. Outgrowth of fibroblast cells from goat skin explants in three different culture media and the establishment of cell lines. In Vitro Cell Dev Biol Anim. 2011;47:83-88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 213. | Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061-1067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 338] [Cited by in F6Publishing: 313] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 214. | Stanton MM, Tzatzalos E, Donne M, Kolundzic N, Helgason I, Ilic D. Prospects for the Use of Induced Pluripotent Stem Cells in Animal Conservation and Environmental Protection. Stem Cells Transl Med. 2019;8:7-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 215. | Verma R, Holland MK, Temple-Smith P, Verma PJ. Inducing pluripotency in somatic cells from the snow leopard (Panthera uncia), an endangered felid. Theriogenology 2012; 77: 220-228, 228.e1-228. e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 216. | Ramaswamy K, Yik WY, Wang XM, Oliphant EN, Lu W, Shibata D, Ryder OA, Hacia JG. Derivation of induced pluripotent stem cells from orangutan skin fibroblasts. BMC Res Notes. 2015;8:577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 217. | Verma R, Liu J, Holland MK, Temple-Smith P, Williamson M, Verma PJ. Nanog is an essential factor for induction of pluripotency in somatic cells from endangered felids. Biores Open Access. 2013;2:72-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 218. | Selvaraj V, Wildt DE, Pukazhenthi BS. Induced pluripotent stem cells for conserving endangered species? Nat Methods. 2011;8:805-807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 219. | Hildebrandt TB, Hermes R, Colleoni S, Diecke S, Holtze S, Renfree MB, Stejskal J, Hayashi K, Drukker M, Loi P, Göritz F, Lazzari G, Galli C. Embryos and embryonic stem cells from the white rhinoceros. Nat Commun. 2018;9:2589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 220. | Fan N, Lai L. Genetically modified pig models for human diseases. J Genet Genomics. 2013;40:67-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 221. | Moradi S, Mahdizadeh H, Šarić T, Kim J, Harati J, Shahsavarani H, Greber B, Moore JB 4th. Research and therapy with induced pluripotent stem cells (iPSCs): social, legal, and ethical considerations. Stem Cell Res Ther. 2019;10:341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 105] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 222. | Sun W, Zheng W, Simeonov A. Drug discovery and development for rare genetic disorders. Am J Med Genet A. 2017;173:2307-2322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 223. | Hannoun Z, Steichen C, Dianat N, Weber A, Dubart-Kupperschmitt A. The potential of induced pluripotent stem cell derived hepatocytes. J Hepatol. 2016;65:182-199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 224. | Bruyneel AA, McKeithan WL, Feyen DA, Mercola M. Will iPSC-cardiomyocytes revolutionize the discovery of drugs for heart disease? Curr Opin Pharmacol. 2018;42:55-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 225. | Rowe RG, Daley GQ. Induced pluripotent stem cells in disease modelling and drug discovery. Nat Rev Genet. 2019;20:377-388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 320] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 226. | Paik DT, Chandy M, Wu JC. Patient and Disease-Specific Induced Pluripotent Stem Cells for Discovery of Personalized Cardiovascular Drugs and Therapeutics. Pharmacol Rev. 2020;72:320-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 99] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 227. | Argentati C, Tortorella I, Bazzucchi M, Morena F, Martino S. Harnessing the Potential of Stem Cells for Disease Modeling: Progress and Promises. J Pers Med. 2020;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 228. | Perleberg C, Kind A, Schnieke A. Genetically engineered pigs as models for human disease. Dis Model Mech. 2018;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 117] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 229. | Malhi PS, Adams GP, Singh J. Bovine model for the study of reproductive aging in women: follicular, luteal, and endocrine characteristics. Biol Reprod. 2005;73:45-53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 115] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 230. | Herath S, Dobson H, Bryant CE, Sheldon IM. Use of the cow as a large animal model of uterine infection and immunity. J Reprod Immunol. 2006;69:13-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 231. | Yapura J, Mapletoft RJ, Pierson R, Singh J, Naile J, Giesy JP, Adams GP. A bovine model for examining the effects of an aromatase inhibitor on ovarian function in women. Fertil Steril 2011; 96: 434-438. e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 232. | Kay MA, Landen CN, Rothenberg SR, Taylor LA, Leland F, Wiehle S, Fang B, Bellinger D, Finegold M, Thompson AR. In vivo hepatic gene therapy: complete albeit transient correction of factor IX deficiency in hemophilia B dogs. Proc Natl Acad Sci USA. 1994;91:2353-2357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 193] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 233. | Wolfe JH. Gene therapy in large animal models of human genetic diseases. Introduction. ILAR J. 2009;50:107-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 234. | Chan AW, Cheng PH, Neumann A, Yang JJ. Reprogramming Huntington monkey skin cells into pluripotent stem cells. Cell Reprogram. 2010;12:509-517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 235. | Camnasio S, Delli Carri A, Lombardo A, Grad I, Mariotti C, Castucci A, Rozell B, Lo Riso P, Castiglioni V, Zuccato C, Rochon C, Takashima Y, Diaferia G, Biunno I, Gellera C, Jaconi M, Smith A, Hovatta O, Naldini L, Di Donato S, Feki A, Cattaneo E. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington's disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis. 2012;46:41-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 136] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 236. | Tu Z, Yang W, Yan S, Guo X, Li XJ. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener. 2015;10:35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 237. | Karagiannis P, Takahashi K, Saito M, Yoshida Y, Okita K, Watanabe A, Inoue H, Yamashita JK, Todani M, Nakagawa M, Osawa M, Yashiro Y, Yamanaka S, Osafune K. Induced Pluripotent Stem Cells and Their Use in Human Models of Disease and Development. Physiol Rev. 2019;99:79-114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 175] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 238. | Tippett P. Blood group chimeras. A review. Vox Sang. 1983;44:333-359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 45] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 239. | Wu J, Greely HT, Jaenisch R, Nakauchi H, Rossant J, Belmonte JC. Stem cells and interspecies chimaeras. Nature. 2016;540:51-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 240. | Mascetti VL, Pedersen RA. Human-Mouse Chimerism Validates Human Stem Cell Pluripotency. Cell Stem Cell. 2016;18:67-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 241. | Tarkowski AK. Mouse chimaeras developed from fused eggs. Nature. 1961;190:857-860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 410] [Cited by in F6Publishing: 422] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 242. | Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Sato H, Lee YS, Usui J, Knisely AS, Hirabayashi M, Nakauchi H. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142:787-799. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 402] [Cited by in F6Publishing: 385] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 243. | Matsunari H, Nagashima H, Watanabe M, Umeyama K, Nakano K, Nagaya M, Kobayashi T, Yamaguchi T, Sumazaki R, Herzenberg LA, Nakauchi H. Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc Natl Acad Sci U S A. 2013;110:4557-4562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 184] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 244. | Yamaguchi T, Sato H, Kato-Itoh M, Goto T, Hara H, Sanbo M, Mizuno N, Kobayashi T, Yanagida A, Umino A, Ota Y, Hamanaka S, Masaki H, Rashid ST, Hirabayashi M, Nakauchi H. Interspecies organogenesis generates autologous functional islets. Nature. 2017;542:191-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 179] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 245. | James D, Noggle SA, Swigut T, Brivanlou AH. Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol. 2006;295:90-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 115] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 246. | Polzin VJ, Anderson DL, Anderson GB, BonDurant RH, Butler JE, Pashen RL, Penedo MC, Rowe JD. Production of sheep-goat chimeras by inner cell mass transplantation. J Anim Sci. 1987;65:325-330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 247. | Tachibana M, Sparman M, Ramsey C, Ma H, Lee HS, Penedo MC, Mitalipov S. Generation of chimeric rhesus monkeys. Cell. 2012;148:285-295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 248. | Bożyk K, Gilecka K, Humięcka M, Szpila M, Suwińska A, Tarkowski AK. Mouse↔rat aggregation chimaeras can develop to adulthood. Dev Biol. 2017;427:106-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 249. | Jaszczak K, Parada R, Guszkiewicz A. Cytogenetic study of some tissues and age-related changes in cell proportions in a goat-sheep chimera. Cytogenet Cell Genet. 1999;84:55-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 250. | Bian G, Qin Q, Feng G, Lu F, Shi D. A preliminary study on making interspecific chimeras between cattle and buffalo by aggregating blastomeres. China Animal Husbandry and Veterinary Medicine. 2007;34:44-46 Available from: http://en.cnki.com.cn/article_en/cjfdtotal-gwxk200709013.htm. [Cited in This Article: ] |
| 251. | Guo J, Wu B, Li S, Bao S, Zhao L, Hu S, Sun W, Su J, Dai Y, Li X. Contribution of Mouse Embryonic Stem Cells and Induced Pluripotent Stem Cells to Chimeras through Injection and Coculture of Embryos. Stem Cells Int. 2014;2014:409021. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 252. | Yang CM, Gong XL, Qiu J, Tang HX, Gong ZJ, Huang SZ, Zeng F. Engraftment of genetically modified human amniotic fluid-derived progenitor cells to produce coagulation factor IX after in utero transplantation in mice. Cell Biol Int. 2013;37:420-429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 253. | Vilarino M, Rashid ST, Suchy FP, McNabb BR, van der Meulen T, Fine EJ, Ahsan SD, Mursaliyev N, Sebastiano V, Diab SS, Huising MO, Nakauchi H, Ross PJ. CRISPR/Cas9 microinjection in oocytes disables pancreas development in sheep. Sci Rep. 2017;7:17472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 254. | Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372-375. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 325] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 255. | Porsdam Mann S, Sun R, Hermerén G. A framework for the ethical assessment of chimeric animal research involving human neural tissue. BMC Med Ethics. 2019;20:10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 256. | Kwisda K, White L, Hübner D. Ethical arguments concerning human-animal chimera research: a systematic review. BMC Med Ethics. 2020;21:24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 257. | Slade P. If you build it, will they eat it? Appetite. 2018;125:428-437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 134] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 258. | Post MJ. Cultured meat from stem cells: challenges and prospects. Meat Sci. 2012;92:297-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 273] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 259. | Scollan ND, Greenwood PL, Newbold CJ, Yanez Ruiz DR, Shingfield KJ, Wallace RJ, Hocquette, JF. Future research priorities for animal production in a changing world. Anim Prod Sci. 2011;51(1):1-5. [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 260. | Arshad MS, Javed M, Sohaib M, Saeed F, Imran A, Amjad Z. Tissue engineering approaches to develop cultured meat from cells: A mini review. Cogent Food & Agri. 2017;3:1320814. [DOI] [Cited in This Article: ] |
| 261. | Genovese NJ, Domeier TL, Telugu BP, Roberts RM. Enhanced Development of Skeletal Myotubes from Porcine Induced Pluripotent Stem Cells. Sci Rep. 2017;7:41833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 262. | Yuan Y. Capturing bovine pluripotency. Proc Natl Acad Sci USA. 2018;115:1962-1963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 263. | Yan X, Zhu MJ, Dodson MV, Du M. Developmental programming of fetal skeletal muscle and adipose tissue development. J Genomics. 2013;1:29-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 264. | Chal J, Pourquié O. Making muscle: skeletal myogenesis in vivo and in vitro. Development. 2017;144:2104-2122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 375] [Cited by in F6Publishing: 449] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 265. | Pimentel D, Pimentel M. Sustainability of meat-based and plant-based diets and the environment. Am J Clin Nutr. 2003;78:660S-663S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 616] [Cited by in F6Publishing: 377] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 266. | Stephens N, Di Silvio L, Dunsford I, Ellis M, Glencross A, Sexton A. Bringing cultured meat to market: Technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci Technol. 2018;78:155-166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 218] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 267. | Bhat ZF, Morton JD, Mason SL, Bekhit AEA. Current and future prospects for the use of pulsed electric field in the meat industry. Crit Rev Food Sci Nutr. 2019;59:1660-1674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 268. | Martin U. Therapeutic Application of Pluripotent Stem Cells: Challenges and Risks. Front Med (Lausanne). 2017;4:229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 269. | Doss MX, Sachinidis A. Current Challenges of iPSC-Based Disease Modeling and Therapeutic Implications. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 234] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 270. | Kusumoto D, Lachmann M, Kunihiro T, Yuasa S, Kishino Y, Kimura M, Katsuki T, Itoh S, Seki T, Fukuda K. Automated Deep Learning-Based System to Identify Endothelial Cells Derived from Induced Pluripotent Stem Cells. Stem Cell Reports. 2018;10:1687-1695. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 271. | Cito M, Pellegrini S, Piemonti L, Sordi V. The potential and challenges of alternative sources of β cells for the cure of type 1 diabetes. Endocr Connect. 2018;7:R114-R125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 27] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 272. | Wu X, Song M, Yang X, Liu X, Liu K, Jiao C, Wang J, Bai C, Su G, Liu X, Li G. Establishment of bovine embryonic stem cells after knockdown of CDX2. Sci Rep. 2016;6:28343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 273. | Verma OP, Kumar R, Nath A, Sharma M, Dubey PK, Kumar GS, Sharma GT. In vivo differentiation potential of buffalo (Bubalus bubalis) embryonic stem cell. In Vitro Cell Dev Biol Anim. 2012;48:349-358. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 274. | Pilichi S, Rocca S, Dattena M, Pool RR, Mara L, Sanna D, Masala G, Manunta ML, Dore S, Manunta A, Passino ES. Sheep embryonic stem-like cells engrafted into sheep femoral condyle osteochondral defects: 4-year follow-up. BMC Vet Res. 2018;14:213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 275. | Zhao Y, Lin J, Wang L, Chen B, Zhou C, Chen T, Guo M, He S, Zhang N, Liu C, Liu M, Huang J. Derivation and characterization of ovine embryonic stem-like cell lines in semi-defined medium without feeder cells. J Exp Zool A Ecol Genet Physiol. 2011;315:639-648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 276. | Cha HJ, Yun JI, Han NR, Kim HY, Baek S, Lee SH, Lee J, Lee E, Park CK, Lee ST. Generation of embryonic stem-like cells from in vivo-derived porcine blastocysts at a low concentration of basic fibroblast growth factor. Reprod Domest Anim. 2018;53:176-185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 277. | Hou DR, Jin Y, Nie XW, Zhang ML, Ta N, Zhao LH, Yang N, Chen Y, Wu ZQ, Jiang HB, Li YR, Sun QY, Dai YF, Li RF. Derivation of Porcine Embryonic Stem-Like Cells from In Vitro-Produced Blastocyst-Stage Embryos. Sci Rep. 2016;6:25838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 278. | Zhang M, Wang C, Jiang H, Liu M, Yang N, Zhao L, Hou D, Jin Y, Chen Q, Chen Y, Wang J, Dai Y, Li R. Derivation of novel naive-like porcine embryonic stem cells by a reprogramming factor-assisted strategy. FASEB J. 2019;33:9350-9361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 279. | Li X, Zhou SG, Imreh MP, Ahrlund-Richter L, Allen WR. Horse embryonic stem cell lines from the proliferation of inner cell mass cells. Stem Cells Dev. 2006;15:523-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 67] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 280. | Abavisani A, Mckinnon AO, Tecirlioglu RT, Trounson AO, Guo J. Maintenance of horse embryonic stem cells in different conditions. Iranian J Vet Res. 2010;3:32. [DOI] [Cited in This Article: ] |
| 281. | Tobias IC, Brooks CR, Teichroeb JH, Villagómez DA, Hess DA, Séguin CA, Betts DH. Small-Molecule Induction of Canine Embryonic Stem Cells Toward Naïve Pluripotency. Stem Cells Dev. 2016;25:1208-1222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 282. | Sandmaier SE, Nandal A, Powell A, Garrett W, Blomberg L, Donovan DM, Talbot N, Telugu BP. Generation of induced pluripotent stem cells from domestic goats. Mol Reprod Dev. 2015;82:709-721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 283. | Chen H, Zuo Q, Wang Y, Song J, Yang H, Zhang Y, Li B. Inducing goat pluripotent stem cells with four transcription factor mRNAs that activate endogenous promoters. BMC Biotechnol. 2017;17:11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 284. | Xu J, Yu L, Guo J, Xiang J, Zheng Z, Gao D, Shi B, Hao H, Jiao D, Zhong L, Wang Y, Wu J, Wei H, Han J. Generation of pig induced pluripotent stem cells using an extended pluripotent stem cell culture system. Stem Cell Res Ther. 2019;10:193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 41] [Article Influence: 8.2] [Reference Citation Analysis (3)] |
| 285. | Setthawong P, Phakdeedindan P, Tiptanavattana N, Rungarunlert S, Techakumphu M, Tharasanit T. Generation of porcine induced-pluripotent stem cells from Sertoli cells. Theriogenology. 2019;127:32-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 286. | Moro LN, Amin G, Furmento V, Waisman A, Garate X, Neiman G, La Greca A, Santín Velazque NL, Luzzani C, Sevlever GE, Vichera G, Miriuka SG. MicroRNA characterization in equine induced pluripotent stem cells. PLoS One. 2018;13:e0207074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 287. | Nishimura T, Hatoya S, Kanegi R, Wijesekera DPH, Sanno K, Tanaka E, Sugiura K, Hiromitsu Tamada NK, Imai H, Inaba T. Feeder-independent canine induced pluripotent stem cells maintained under serum-free conditions. Mol Reprod Dev. 2017;84:329-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 288. | Gonçalves NJN, Bressan FF, Roballo KCS, Meirelles FV, Xavier PLP, Fukumasu H, Williams C, Breen M, Koh S, Sper R, Piedrahita J, Ambrósio CE. Generation of LIF-independent induced pluripotent stem cells from canine fetal fibroblasts. Theriogenology. 2017;92:75-82. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |