Published online Jul 7, 2019. doi: 10.3748/wjg.v25.i25.3116
Peer-review started: February 22, 2019
First decision: April 30, 2019
Revised: May 14, 2019
Accepted: May 31, 2019
Article in press: June 1, 2019
Published online: July 7, 2019
The intestinal epithelium possesses a remarkable ability for both proliferation and regeneration. The last two decades have generated major advances in our understanding of the stem cell populations responsible for its maintenance during homeostasis and more recently the events that occur during injury induced regeneration. These fundamental discoveries have capitalised on the use of transgenic mouse models and in vivo lineage tracing to make their conclusions. It is evident that maintenance is driven by rapidly proliferating crypt base stem cells, but complexities associated with the technicality of mouse modelling have led to several overlapping populations being held responsible for the same behaviour. Similarly, it has been shown that essentially any population in the intestinal crypt can revert to a stem cell state given the correct stimulus during epithelial regeneration. Whilst these observations are profound it is uncertain how relevant they are to human intestinal homeostasis and pathology. Here, these recent studies are presented, in context with technical considerations of the models used, to argue that their conclusions may indeed not be applicable in understanding “homeostatic regeneration” and experimental suggestions presented for validating their results in human tissue.
Core tip: Recent advances, using transgenic mice, in understanding cellular hierarchies in the intestinal epithelium have identified numerous cell populations which retain the ability to change their fate in response to injury. Here, these new studies are presented in the context of a discussion about what represents a relevant epithelial injury to understand “homeostatic regeneration”. Experimental suggestions are proposed for validating animal findings to translate our current knowledge to better understand human intestinal epithelial maintenance.
- Citation: Buczacki S. Fate plasticity in the intestine: The devil is in the detail. World J Gastroenterol 2019; 25(25): 3116-3122
- URL: https://www.wjgnet.com/1007-9327/full/v25/i25/3116.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i25.3116
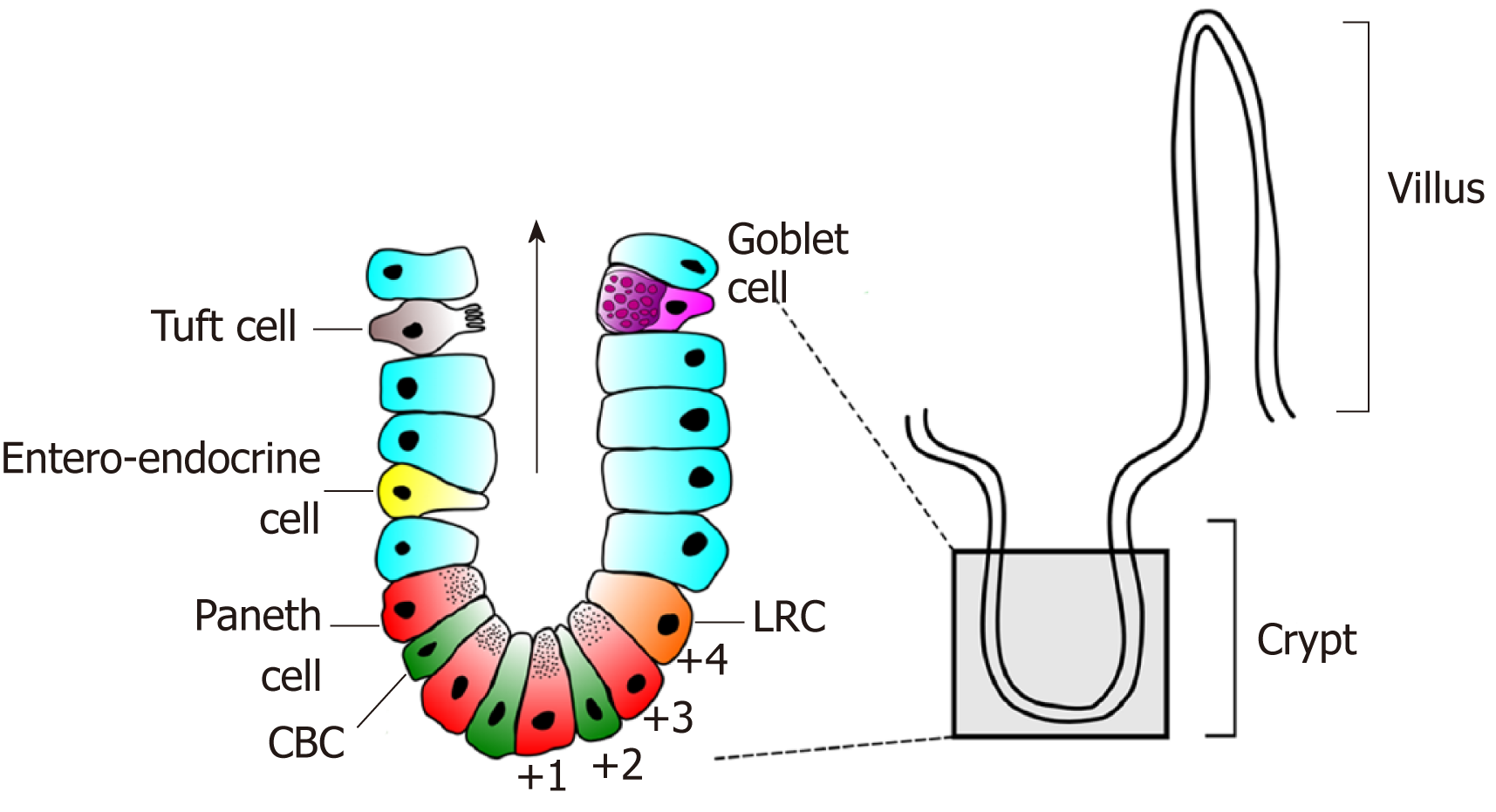
The intestinal lining is one of the most rapidly proliferating epithelia in humans. In the small intestine this single-cell thick structure is thrown into folds consisting of villi that protrude into the lumen and crypts that are embedded in the intestinal wall. The colonic epithelium is similar although lacks villi. At a histological level the small intestine and colon are also alike. In the small intestine the most prevalent cell type is the absorptive enterocyte and, in the colon, the colonocyte. Both organs also possess secretory goblet, enteroendocrine and tuft cells (Figure 1). The main cellular difference between the small intestine and colon is the presence of secretory Paneth cells. These long-lived secretory and niche cells are only found in the bottom of small intestinal crypts and rarely found in the normal colon. A functionally similar cell type, termed the deep crypt secretory cell, has however recently been shown to exist in colonic crypts[1,2]. Both epithelia are highly proliferative and retain a remarkable ability for regeneration following injury. Homeostatic proliferation throughout the intestine takes place in the bottom of the crypts, being most active in the so-called transit amplifying zone – an area directly above the crypt base. Differentiation occurs as cells migrate up the crypts onto either villi or the colonic mucosal plateau.
Over the last two decades the intestine has become an area of great interest in stem cell biology and is arguably the prototypical organ for the study of epithelial homeostasis and regeneration partly due to the unique structure of the crypt that facilitates ready quantification of stem cell clonogenicity. Following seminal findings using lineage tracing, of the clonogenic function of undifferentiated crypt cells during homeostasis, the concept of cellular plasticity has more recently been explored using contemporary in vivo techniques. Plasticity is defined as a change in cell fate in response to a stimulus. The results of these new studies have however led the field into a complex and confusing period where, on face value, it appears that almost any cell type in the intestinal epithelium can revert to a stem cell state during regeneration. In this opinion review I discuss both the important original and more recent studies and propose that whilst the findings are striking they may not be entirely relevant for our understanding of “homeostatic regeneration”. Here, I define homeostatic regeneration as the cellular changes that occur during the response to injury classically occurring during mammalian life and commonly encountered pathologies.
Evidence for the existence of intestinal stem cells was first demonstrated in the 1970s by Cheng and Leblond who showed that after treating mice with tritiated thymidine, crypt base columnar cells (CBCs) developed labelled phagosomes following phagocytosis of nearby non-viable cells[3]. Subsequent tracing of these labelled phagosomes over time found they were inherited by all the differentiated cell types of the epithelium. These experiments proposed that stem cells present in the base of intestinal crypts could generate all the differentiated cell types of the intestinal epithelium. Following this, attention focussed on cells in the so-called +4 position that appeared both quiescent and undifferentiated – a feature commonly found in stem cells in other organs[4]. The field however underwent a sea change in 2007 following the publication from Hans Clevers’ laboratory demonstrating that Lgr5 expression marked rapidly proliferating CBCs in the small intestine and colon which were capable of profound clonal capacity as shown using a lineage tracing technique in mice[5]. This highly elegant study provided the first direct proof that rapidly cycling Lgr5+ CBCs were the bona fide homeostatic stem cells of throughout the intestine. There then followed a period of intense debate about the nature of the +4 cell with several groups showing marker overlay of genes of interest with cells in this position also possessing stem cell capacity including Bmi1, Hopx and mTert[6-8]. Interestingly, like the original Barker et al[5] study, all three of these studies primarily used the location of promoter driven reporter expression to define the anatomical location of cells expressing the respective gene of interest. It is however unknown what degree of gene activity is required to drive reporter expression and this can be compounded by the introduction of Cre recombinase as a conditional activator of reporter expression as is often used in lineage tracing studies. Highlighting these issues, two separate studies of Bmi1 expressing cells using different models; Bmi1-CreER and Bmi1-GFP (both knocked in at the endogenous locus) show different results with one study finding the cells to be stem cells and the other mature enteroendocrine cells[6,9]. The Cre enzyme is also seen to possess apparent regional differences in expression when under the control of reportedly pan intestinal promoters; Cre is often found to have greater activity the more proximal in the intestinal tract making it hard to compare with stem cell behaviour in the distal small intestine and colon[10,11]. Whether this is due to promoter, enzyme intrinsic or reporter differences is incompletely understood. These concerns can be compounded when a CreER system is used to drive conditional recombination. In this situation off-target effects of both tamoxifen and impaired stem cell function following activation of Cre have been reported by two separate studies[12,13]. These important studies indicate that quantification of stem cell behaviour following tamoxifen driven Cre activation may not be accurate or representative of the true in vivo situation.
Tissue specific gene promoters can also have problems with both sensitivity and specificity for all cells on the crypt-villus axis. A comparison between two intestinal, reportedly pan-epithelial Cre models, Villin-Cre and Ah-Cre showed unexpected variation in the Ah-Cre driven recombination that failed to target a cell population that was capable of driving regeneration[14]. Previously the Ah-Cre model had been reported to induce recombination in all IECs other than terminally differentiated Paneth cells[10]. However, in the study by Parry et al[14] the authors found that Ah-Cre also failed to induce recombination in a non-Paneth cell putative reserve stem cell population. The implication of this work is that quantifying global clonal output using Ah-Cre driven reporters may not include all potentially clonogenic populations and erroneous conclusions could be drawn.
It is also unknown how long reporter proteins persist and are visible for, following cessation of their production. It is likely that different reporters have varying stability that could confound analysis if induced conditionally and thus temporally where expression may still be visualised in daughter cells that aren’t expressing the mRNA of the gene of interest. This may lead to an assumption that reporter expression is directly correlated with gene expression which has not definitively be proven. Cells may also express low levels of the gene of interest and still possess the same functionality as those with high levels of expression (as the marker itself is unlikely to directly drive function) but low expressors may fail to be identified by reporter expression. Indeed, precisely these concerns were found valid when bulk gene expression comparisons were used to compare the transcriptome of CBC Lgr5+ cells with +4 located Bmi1+ cells suggesting that these cells are in reality one and the same and the +4 cell may well be Lgr5 expressing[15]. Cumulatively, all these early studies certainly provide direct evidence that homeostatic stem cells exist at the base of the intestinal crypts but also demonstrate the clear difficulties in using transgenic mouse models to dissect subpopulations and functionality at a high level of detail.
Given the striking ability of the intestinal epithelium to regenerate, a natural progression of recent research focus has been to understand which cell populations are responsible for this behaviour. Theoretically there are two possible cell types responsible for regeneration; a distinct quiescent sub-population waiting to become activated when required or a population that has one role during homeostasis but that can revert to a stem-like state during injury i.e., plastic. The first named population found to have a role in regeneration were Dll1 expressing crypt cells[16]. Using a combination of lineage tracing in combination with irradiation the Clevers group found that during homeostasis these cells were proliferating secretory progenitors but following irradiation and cell death they acquired stem cell capacity and were thus capable of regenerating the injured epithelium. The question of what the apparently quiescent +4 cell represented was further addressed by Doug Winton’s group using a novel split-Cre mouse to conditionally genetically mark label-retaining cells (LRCs)[17]. Supporting the findings of the Clevers group this study found LRCs to be a slowly-cycling Paneth and enteroendocrine cell progenitor that similarly reverted to a stem cell-like fate during injury induced regeneration albeit at very low frequency - partly due to the complexities of the transgenic model employed.
Following the finding of plasticity in Dll1 cells and LRCs, attention has focussed on whether other populations can perform similar functions. Indeed, it has now been shown that goblet cells, enteroendocrine cells, enterocytes and Paneth cells are all capable of contextually acquiring stem cell capacity[9,18-28]. Analysis of these reports however shows wide variation in the types of injury models employed varying from relatively mild oral dextran sulfate sodium (DSS) that induces mucosal inflammation, to lethal whole-body irradiation (12Gy), making it difficult to compare results between studies (Table 1). More recently it has also been shown that some secretory progenitor populations even during homeostasis may stochastically acquire stemness[22,26,28]. This bi-fated character of some secretory progenitors was originally demonstrated in 2004 where a small number of Ngn3 enteroendocrine cell progenitors were also found to have clonal/stem cell capacity during homeostasis[29]. Whilst it is entirely plausible that there are a wide range of cell types capable of plasticity there are evidently those that cannot, as clone formation has never been found arising on villi even during classical injury induced regeneration. Schwitalla et al[30] have however demonstrated that aberrant elevated NF-kB signalling in apparently terminally differentiated enterocytes on the villi can cause dedifferentiation to a tumour-initiating Lgr5+ status. This finding proposes that at the very least, if given a strong enough stimulus, even terminally differentiated villus-based enterocytes may acquire some stem cell characteristics.
| Study | Plastic cell identified | Injury model used |
| Tian et al[18] | Bmi1+ cell | DTR Lgr5+ ablation |
| Roth et al[19] | Paneth cell | 12Gy radiation |
| van Es et al[16] | Dll1+ cell | 6Gy radiation |
| Buczacki et al[17] | Label-retaining cell | 6Gy radiation, doxorubicin or hydroxyurea |
| Asfaha et al[20] | Upper crypt progenitor | 12Gy radiation +/- 5-Fluorouracil |
| Tetteh et al[27] | Alpi1+ enterocyte | DTR Lgr5+ ablation |
| Jadhav et al[9] | Goblet cell progenitors | DTR Lgr5+ ablation |
| Yan et al[21] | Enteroendocrine cell | 12Gy radiation |
| Ishibashi et al[22] | Atoh1+ cell | DSS (1.75%) for 5 d |
| Nusse et al[23] | Lgr5- crypt cell | Parasite infection |
| Schmitt et al[24] | Paneth cell | DSS (3%) for 1 wk |
| Tomic et al[26] | Atoh1+ cell | 6Gy radiation, AOM or 2% DSS |
| Yu et al[25] | Lyz1+ Paneth cell | 12Gy radiation |
| Castillo et al[28] | Atoh1+ cell | DSS (2.5%-3%) for 5 d |
Cumulatively, these studies show widespread plasticity amongst almost every cell type described to-date in the murine intestinal epithelium however there are many inconsistencies between studies driven primarily from the technical issues related to ascribing identity and plasticity (Table 2). It could also be argued that the apparent broadly found plasticity may not be relevant to advancing our understanding of what cell types are actually at play in humans during routine epithelial insult. It has been known for decades that cell types can be reprogrammed to different identities and this forms the fundamental basis of induced pluripotent stem cells (iPS) technology. In Waddington’s classical model of the epigenetic landscape of differentiation it is therefore clear that cells can traverse between several deep valleys given the appropriate stimulus to ascend the elevations between[31]. Evidently, any cell type if pushed hard enough can de-differentiate or trans-differentiate but the question remains what represents a physiological injury and what cell types are involved in the subsequent “homeostatic regeneration”? It may well be the case that during different forms of injury such as that seen between Crohn’s disease and ulcerative colitis varying cell types are mobilised via plasticity to the stem cell state to try and repair the damaged epithelium.
| CreER and Tamoxifen | Toxicity |
| Off-target effects | |
| Incongruity between reporter expression and protein expression | Regional differences |
| Chronicity of reporter stability | |
| Reporter and mRNA expression differences | |
| Inconsistent injury models | Intestinal specific effects including incomplete cell type eradication e.g., diphtheria toxin mediated cell death |
| Off-target whole body effects e.g., irradiation | |
| Representative of “homeostatic regeneration” | |
| Different cell-type responses to different injuries | |
| Laboratory differences | Microbiota |
| Diet | |
| Area of intestine examined | |
| Strain differences between laboratories due to inbreeding |
How then can the field move forward to make meaningful in-roads into translating these previous animal findings to the benefit of patients? There are two areas that would seem ripe for development – tools for lineage tracing in humans and better validated injury models in mice. Lineage tracing in mice is primarily performed through generating transgenic mouse strains which is clearly impossible in humans. There are however several new tools that could permit lineage tracing analysis in human tissue albeit by quantifying the clonal output of all potentially clonogenic cells. Two important recent reports have made use of next generation sequencing technology to identify clones through the quantification of somatically acquired mosaic mutations found in human and murine tissue[32,33]. The advances now made in sequencing technologies enabling combined single cell DNA (scDNAseq) and RNA sequencing (scRNAseq) allow for similar sequencing approaches with higher coverage to uncover cellular hierarchies from human tissue in both disease and homeostasis.
Clonal marking in humans can also be performed by quantifying mitochondrial DNA (mtDNA) mutations through dual immunohistochemical (IHC) staining for the mitochondrially encoded enzyme cytochrome-c oxidase (CCO)[34]. Cells acquire mtDNA mutations infrequently and can be identified through this IHC technique as cumulatively more are acquired in the many mtDNA copies of the CCO gene via stochastic genetic drift. As the mutation is genetic and therefore heritable, the clonal output of cells acquiring these mutations can be quantified. More recently several new similarly working but genomic DNA (gDNA) encoded neutral IHC clonal marks have been described by the Winton laboratory[35]. Importantly, both CCO staining and the newer clonal marks allow the quantification of clonal behaviour in situ from human tissue. Using these techniques on matched sections of diseased and normal tissue allows an understanding, at the numerical level, of changes in behaviour such as seen in plasticity. Further, combining these lineage tracing approaches with scRNAseq could provide profound insights, like those described in the mouse, to understanding intestinal homeostasis and plasticity in humans.
Finally, there is an urgent need to better define the cellular effects of common human intestinal epithelial injuries to identify appropriate murine model equivalents. The current use of multiple different forms of injury models which only bear a passing relationship to human disease in that there is some degree of cell death is far from ideal. The interplay between epithelial loss, stromal tissue, immune cells, vasculature and resident microbiota is highly complex and very likely inadequately modelled in our current simplistic models of injury performed on mice housed in clean animal facilities. Here, the opposite approach to that presented earlier could be used: Human studies into the precise cellular events occurring during various injuries could inform the development of better murine injury models. It would appear that the field is reaching the limitations of what can be achieved with current tools and models and in order to advance as rapidly as previously, new approaches are required that maximise on novel technologies and translationally relevant models.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United Kingdom
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Vagholkar KR S-Editor: Ma RY L-Editor: A E-Editor: Ma YJ
| 1. | Rothenberg ME, Nusse Y, Kalisky T, Lee JJ, Dalerba P, Scheeren F, Lobo N, Kulkarni S, Sim S, Qian D, Beachy PA, Pasricha PJ, Quake SR, Clarke MF. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology. 2012;142:1195-1205.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 187] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 2. | Sasaki N, Sachs N, Wiebrands K, Ellenbroek SI, Fumagalli A, Lyubimova A, Begthel H, van den Born M, van Es JH, Karthaus WR, Li VS, López-Iglesias C, Peters PJ, van Rheenen J, van Oudenaarden A, Clevers H. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci USA. 2016;113:E5399-E5407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 191] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 3. | Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537-561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1049] [Cited by in F6Publishing: 1005] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 4. | Potten CS. Extreme sensitivity of some intestinal crypt cells to X and gamma irradiation. Nature. 1977;269:518-521. [PubMed] [Cited in This Article: ] |
| 5. | Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003-1007. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3854] [Cited by in F6Publishing: 4012] [Article Influence: 236.0] [Reference Citation Analysis (0)] |
| 6. | Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915-920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 928] [Cited by in F6Publishing: 925] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 7. | Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci USA. 2011;108:179-184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 392] [Cited by in F6Publishing: 397] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 8. | Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420-1424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 551] [Cited by in F6Publishing: 558] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 9. | Jadhav U, Saxena M, O'Neill NK, Saadatpour A, Yuan GC, Herbert Z, Murata K, Shivdasani RA. Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell Stem Cell. 2017;21:65-77.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 159] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 10. | Ireland H, Kemp R, Houghton C, Howard L, Clarke AR, Sansom OJ, Winton DJ. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology. 2004;126:1236-1246. [PubMed] [Cited in This Article: ] |
| 11. | el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 738] [Cited by in F6Publishing: 799] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 12. | Zhu Y, Huang YF, Kek C, Bulavin DV. Apoptosis differently affects lineage tracing of Lgr5 and Bmi1 intestinal stem cell populations. Cell Stem Cell. 2013;12:298-303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 13. | Bohin N, Carlson EA, Samuelson LC. Genome Toxicity and Impaired Stem Cell Function after Conditional Activation of CreERT2 in the Intestine. Stem Cell Reports. 2018;11:1337-1346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 14. | Parry L, Young M, El Marjou F, Clarke AR. Evidence for a crucial role of paneth cells in mediating the intestinal response to injury. Stem Cells. 2013;31:776-785. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, Myant K, Versteeg R, Sansom OJ, van Es JH, Barker N, van Oudenaarden A, Mohammed S, Heck AJ, Clevers H. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 2012;31:3079-3091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 609] [Cited by in F6Publishing: 562] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 16. | van Es JH, Sato T, van de Wetering M, Lyubimova A, Yee Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, Martens ACM, Barker N, van Oudenaarden A, Clevers H. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 556] [Cited by in F6Publishing: 552] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 17. | Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65-69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 614] [Cited by in F6Publishing: 565] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 18. | Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255-259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 836] [Cited by in F6Publishing: 869] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 19. | Roth S, Franken P, Sacchetti A, Kremer A, Anderson K, Sansom O, Fodde R. Paneth cells in intestinal homeostasis and tissue injury. PLoS One. 2012;7:e38965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 20. | Asfaha S, Hayakawa Y, Muley A, Stokes S, Graham TA, Ericksen RE, Westphalen CB, von Burstin J, Mastracci TL, Worthley DL, Guha C, Quante M, Rustgi AK, Wang TC. Krt19(+)/Lgr5(-) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell Stem Cell. 2015;16:627-638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 146] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 21. | Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A, Roelf K, Calderon RI, Cynn E, Hu X, Mandleywala K, Wilhelmy J, Grimes SM, Corney DC, Boutet SC, Terry JM, Belgrader P, Ziraldo SB, Mikkelsen TS, Wang F, von Furstenberg RJ, Smith NR, Chandrakesan P, May R, Chrissy MAS, Jain R, Cartwright CA, Niland JC, Hong YK, Carrington J, Breault DT, Epstein J, Houchen CW, Lynch JP, Martin MG, Plevritis SK, Curtis C, Ji HP, Li L, Henning SJ, Wong MH, Kuo CJ. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell. 2017;21:78-90.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 224] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 22. | Ishibashi F, Shimizu H, Nakata T, Fujii S, Suzuki K, Kawamoto A, Anzai S, Kuno R, Nagata S, Ito G, Murano T, Mizutani T, Oshima S, Tsuchiya K, Nakamura T, Watanabe M, Okamoto R. Contribution of ATOH1+ Cells to the Homeostasis, Repair, and Tumorigenesis of the Colonic Epithelium. Stem Cell Reports. 2018;10:27-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 23. | Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, de Sauvage FJ, Locksley RM, Klein OD. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature. 2018;559:109-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 180] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 24. | Schmitt M, Schewe M, Sacchetti A, Feijtel D, van de Geer WS, Teeuwssen M, Sleddens HF, Joosten R, van Royen ME, van de Werken HJG, van Es J, Clevers H, Fodde R. Paneth Cells Respond to Inflammation and Contribute to Tissue Regeneration by Acquiring Stem-like Features through SCF/c-Kit Signaling. Cell Rep. 2018;24:2312-2328.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 139] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 25. | Yu S, Tong K, Zhao Y, Balasubramanian I, Yap GS, Ferraris RP, Bonder EM, Verzi MP, Gao N. Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell. 2018;23:46-59.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 26. | Tomic G, Morrissey E, Kozar S, Ben-Moshe S, Hoyle A, Azzarelli R, Kemp R, Chilamakuri CSR, Itzkovitz S, Philpott A, Winton DJ. Phospho-regulation of ATOH1 Is Required for Plasticity of Secretory Progenitors and Tissue Regeneration. Cell Stem Cell. 2018;23:436-443.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 27. | Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, van Oudenaarden A, Clevers H. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell. 2016;18:203-213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 373] [Cited by in F6Publishing: 380] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 28. | Castillo-Azofeifa D, Fazio EN, Nattiv R, Good HJ, Wald T, Pest MA, de Sauvage FJ, Klein OD, Asfaha S. Atoh1+ secretory progenitors possess renewal capacity independent of Lgr5+ cells during colonic regeneration. EMBO J. 2019;38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 29. | Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol. 2004;270:443-454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 223] [Cited by in F6Publishing: 221] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 30. | Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, Rupec RA, Gerhard M, Schmid R, Barker N, Clevers H, Lang R, Neumann J, Kirchner T, Taketo MM, van den Brink GR, Sansom OJ, Arkan MC, Greten FR. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 821] [Cited by in F6Publishing: 772] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 31. | Waddington CH. Organisers and Genes. Cambridge: Cambridge Univ. Press 1940; . [Cited in This Article: ] |
| 32. | Behjati S, Huch M, van Boxtel R, Karthaus W, Wedge DC, Tamuri AU, Martincorena I, Petljak M, Alexandrov LB, Gundem G, Tarpey PS, Roerink S, Blokker J, Maddison M, Mudie L, Robinson B, Nik-Zainal S, Campbell P, Goldman N, van de Wetering M, Cuppen E, Clevers H, Stratton MR. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422-425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 267] [Cited by in F6Publishing: 252] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 33. | Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, Huch M, Boymans S, Kuijk E, Prins P, Nijman IJ, Martincorena I, Mokry M, Wiegerinck CL, Middendorp S, Sato T, Schwank G, Nieuwenhuis EE, Verstegen MM, van der Laan LJ, de Jonge J, IJzermans JN, Vries RG, van de Wetering M, Stratton MR, Clevers H, Cuppen E, van Boxtel R. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 718] [Cited by in F6Publishing: 605] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 34. | Walther V, Alison MR. Cell lineage tracing in human epithelial tissues using mitochondrial DNA mutations as clonal markers. Wiley Interdiscip Rev Dev Biol. 2016;5:103-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 35. | Nicholson AM, Olpe C, Hoyle A, Thorsen AS, Rus T, Colombé M, Brunton-Sim R, Kemp R, Marks K, Quirke P, Malhotra S, Ten Hoopen R, Ibrahim A, Lindskog C, Myers MB, Parsons B, Tavaré S, Wilkinson M, Morrissey E, Winton DJ. Fixation and Spread of Somatic Mutations in Adult Human Colonic Epithelium. Cell Stem Cell. 2018;22:909-918.e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |