Published online Oct 21, 2015. doi: 10.3748/wjg.v21.i39.11141
Peer-review started: March 11, 2015
First decision: April 24, 2015
Revised: May 25, 2015
Accepted: August 29, 2015
Article in press: August 29, 2015
Published online: October 21, 2015
AIM: To correlate a genetic polymorphism of the low-density lipoprotein (LDL) receptor with antiviral responses in Egyptian chronic hepatitis C virus (HCV) patients.
METHODS: Our study included 657 HCV-infected patients with genotype 4 who received interferon-based combination therapy. Patients were divided into two groups based on their response to therapy: 356 were responders, and 301 were non-responders. Patients were compared to 160 healthy controls. All patients and controls underwent a thorough physical examination, measurement of body mass index (BMI) and the following laboratory tests: serum alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, albumin, total bilirubin, direct bilirubin, prothrombin time, prothrombin concentration, INR, complete blood count, serum creatinine, fasting blood sugar, HCV antibody, and hepatitis B surface antigen. All HCV patients were further subjected to the following laboratory tests: HCV-RNA using quantitative polymerase chain reaction (PCR), antinuclear antibodies, thyroid-stimulating hormone, an LDL receptor (LDLR) genotype study of LDLR exon8c.1171G>A and exon10c.1413G>A using real-time PCR-based assays, abdominal ultrasonography, ultrasonographic-guided liver biopsy, and histopathological examination of liver biopsies. Correlations of LDL receptor polymorphisms with HAI, METAVIR score, presence of steatosis, and BMI were performed in all cases.
RESULTS: There were no statistically significant differences in response rates between the different types of interferon used or LDLR exon10c.1413G>A. However, there was a significant difference in the frequency of the LDL receptor exon8c.1171G>A genotype between cases (AA: 25.9%, GA: 22.2%, GG: 51.9%) and controls (AA: 3.8%, GA: 53.1% and GG: 43.1%) (P < 0.001). There was a statistically significant difference in the frequency of the LDLR exon 8C:1171 G>A polymorphism between responders (AA: 3.6%, GA: 15.2%, GG: 81.2%) and non-responders (AA: 52.2%, GA: 30.6%, GG: 17.2%) (P < 0.001). The G allele of LDL receptor exon8c.1171G>A predominated in cases and controls over the A allele, and a statistically significant association with response to interferon was observed. The frequency of the LDLR exon8c.1171G>A allele in non-responders was: A: 67.4% and G: 32.6 vs A: 11.2% and G: 88.8% in responders (P < 0.001). Therefore, carriers of the A allele exhibited a 16.4 times greater risk for non-response. There was a significant association between LDL receptors exon8 c.1171G>A and HAI (P < 0.011). There was a significant association between LDL receptors exon8c.1171G>A and BMI. The mean BMI level was highest in patients carrying the AA genotype (28.7 ± 4.7 kg/m2) followed by the GA genotype (28.1 ± 4.8 kg/m2). The lowest BMI was the GG genotype (26.6 ± 4.3 kg/m2) (P < 0.001). The only significant associations were found between LDL receptors exon8 c.1171G>A and METAVIR score or steatosis (P < 0.001).
CONCLUSION: LDL receptor gene polymorphisms play a role in the treatment response of HCV and the modulation of disease progression in Egyptians infected with chronic HCV.
Core tip: Two molecules may function as hepatitis C virus (HCV) receptors, namely the low density lipoprotein receptor (LDLR) and CD81. This work assessed the role of genetic polymorphisms of the LDLR in Egyptian chronic HCV patients and its correlation with antiviral responses to treatment with pegylated interferon /ribavirin therapy. The study demonstrated that LDLR gene polymorphisms play a role in the response to viral treatment. The G allele of LDLRs exon8c.1171G>A predominated in cases and controls over the A allele. Carriers of the A allele exhibited a 16.4 times greater risk for non-response.
- Citation: Naga M, Amin M, Algendy D, Elbadry A, Fawzi M, Foda A, Esmat S, Sabry D, Rashed L, Gabal S, Kamal M. Low-density lipoprotein receptor genetic polymorphism in chronic hepatitis C virus Egyptian patients affects treatment response. World J Gastroenterol 2015; 21(39): 11141-11151
- URL: https://www.wjgnet.com/1007-9327/full/v21/i39/11141.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i39.11141
Hepatitis C exhibits a worldwide prevalence of approximately 3%[1]. Hepatitis C causes severe morbidity and mortality, and the majority of infected subjects fail to eliminate the virus and develop chronic hepatitis C. Chronic infections are estimated at approximately 80% of infected subjects, and this chronicity will lead to cirrhosis and /or hepatocellular carcinoma[2-8].
Egypt has the largest epidemic of hepatitis C virus (HCV) worldwide with a percentage of 14.7%, which is ten times greater than any other country[9-14].
The strong prevalence of HCV subtype (4a) in Egypt suggests that this chronic infection began as an epidemic. A history of injection treatment was implicated as a risk factor for HCV, and the prime culprit is the past practice of parental therapy for schistosomiasis[15,16].
The rate of disease progression varies widely, and it is not known what factors precisely determine the clinical outcome of the disease in the long term. The mechanism of viral entry into the host hepatocytes is not known.
Much effort was made to identify the receptors involved in viral entry into host cells.
Two molecules were proposed to function as HCV receptors, namely the low density lipoprotein receptors (LDLRs) and CD81[17-20].
The LDLR gene family functions as a receptor for the minor group of common cold viruses and other viruses[21,22].
LDLR plays an important role in cholesterol homeostasis[23,24], and it is firmly established that mutations and polymorphisms in the LDLR gene are associated with familial hypercholesterolemia, obesity and atherosclerosis[25-31].
A direct interaction of HCV E2 envelope protein and LDL mediates the binding and internalisation of lipoviroparticles by LDLR in conjunction with CD81[32]. LDLR, CD81 and HCV E2 are co-localized on cells[33], and viral binding and endocytosis are inhibited by free LDL, free recombinant LDLR peptides, and antibodies against E2 and LDLRs[32,34]. Moreover, intracellular HCV RNA levels positively correlate with LDLR expression in cultured primary hepatocytes and in vivo[34,35]. These findings demonstrate that LDLRs are crucial co-receptors for HCV cell entry and replication. Furthermore, the role of LDLRs in immune responses and the incrimination of LDLR polymorphisms in disease processes makes these receptors an important candidate for the study of the genetic susceptibility to hepatitis C.
Several studies investigated nine single nucleotide polymorphisms in the LDLR gene and their possible influence on clinical parameters of HCV infection, such as viral clearance, overall inflammation, fibrosis severity and treatment response[36-38].
Our study assessed genetic polymorphisms of LDLR in chronic HCV genotype 4 Egyptian patients and the effects of polymorphisms on antiviral response.
The review board of the Department of Internal Medicine, Faculty of Medicine, Cairo University approved the study protocol, which was performed according to the Declaration of Helsinki.
All study participants provided informed written consent prior to study enrolment. The study included 657 chronic HCV patients who were candidates for pegylated-interferon/ribavirin therapy and 160 healthy age- and sex-matched subjects as a control group. Patients were recruited from the outpatient clinics of the Internal Medicine Department, Kasr Al Aini Hospital, Cairo University, Beni-Suef Public Hospital and the National Liver Institute.
All patients and control group underwent thorough physical examinations, measurements of body mass index (BMI) and the following laboratory tests: serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), albumin, total bilirubin, direct bilirubin, prothrombin time (PT), prothrombin concentration (PC), INR, complete blood count (CBC), serum creatinine, fasting blood sugar (FBS), HCV antibody (anti-HCV), and hepatitis B surface antigen (HBsAg).
All HCV patients were further subjected to the following tests: HCV-RNA using quantitative polymerase chain reaction (PCR), antinuclear antibodies (ANA), thyroid-stimulating hormone (TSH), LDLR genotype study, abdominal ultrasonography and ultrasonographic-guided liver biopsy.
Patients received a course of antiviral therapy that consisted of pegylated interferon subcutaneously once weekly and oral ribavirin (10.6 mg/kg) daily. Patients with a reduction of more than 2 logs in PCR results after 12 wk of treatment were deemed responders, and treatment was continued for a total of 48 wk. Patients with PCR scores that were not at least 2 logs lower than baseline after 12 wk of therapy were deemed non-responders. Responders were re-tested 6 mo after the end of therapy using PCR to ensure a sustained virological response (SVR). We divided the patients into two groups based on treatment response: non-responders (n = 301) and responders (n = 356).
The following types of pegylated interferon were used based on availability: Peginterferon alfa-2a (PEGASYS® 180 mcg) was used in 287 cases, Peginterferon alfa-2a (Reiferon Retard® 160 mcg) was used in 59 cases, and Peginterferon alfa-2b (PEGINTRON® 1.5 mcg/kg per week) was used in 311 cases.
Venous blood (10 mL) was collected from each patient in EDTA vacuum whole blood sample tubes. Samples were divided into 2 parts: 5 mL for HCV and biochemical parameters assessments and the other 5 mL for molecular LDLR gene polymorphism assessments.
AST, ALT, T. bilirubin, D. bilirubin, albumin, ALP, FBS and creatinine were assessed using commercially available kits (Randox Laboratories Limited, Country Antrim, United Kingdom). AFP was evaluated using an ELISA kit (DRG International Inc., Springfield, New Jersey, United States). HbA 1c, PC, PT and INR were detected using kits (Stanbio Laboratory, Boerne, TX, United States). HB, TLC, ANC and platelets were detected using a cell counter (Sysmex XT-4000i Automated Hematology Analyzer Lincolnshire, IL, United States).
HCV RNA was extracted from 140 μL serum using the QIAamp Viral RNA Mini Kit (QIAgen, Hilden, Germany). Absolute quantitation of the concentration of HCV RNA was based on an external standard curve (HCV Standards IU/mL) in the presence of an internal positive control (IPC). IPC was added to a mixture of lysis buffer and sample material during RNA extraction of clinical blood samples. TaqMan assay was used in the AgPath-IDTM One-Step RT-PCR kit (Applied Biosystems, Foster City, CA, United States). The One-Step RT-PCR kit included an enzyme mixture, buffer and detection enhancer for one-step quantitative reverse transcription PCR (qRT-PCR). Amplification was performed in 25 μL of reaction mixture containing 2 × TaqMan Universal RT-PCR Master Mix, 20 μmol/L of each primer and probes for the sample and ICP and 8.5 μL of extracted RNA. All samples were performed in duplicate. Amplification began with an incubation at 50 °C for two min with uracil N’-glycosylase to inactivate possible contaminating amplicons, followed by 45 °C for 10 min for cDNA synthesis using reverse transcriptase, and 10 min at 95 °C to activate the AmpliTaq Gold polymerase and inactivate uracil N’-glycosylase. The PCR cycling program consisted of 45 cycles of 15 s at 95 °C and 45 s at 60 °C (universal conditions).
Total DNA was isolated from whole blood mononuclear cells (MNCs) using an extraction kit (Qiagene, United States) according to manufacturer’s instructions. DNA purity (A260/A280 ratio) and concentration were calculated using spectrophotometry (dual wavelength Beckman, Spectrophotometer, United States). The extracted and purified DNA samples were stored at -80 °C for further use.
LDLR allelic discrimination variants were genotyped using real-time PCR and the QuantiTect® Probe PCR Kit (Qiagen, Hilden, Germany) and a Step-One instrument (Applied Biosystems, Foster City, United States). PCR products were generated under standard conditions in a 15 μL reaction containing 1 × reaction buffer, 1-2 mmol/L MgCl2, 0.32 mmol/L dNTPs, 0.12 μmol/L of forward, reverse primer and probes, 1 unit Taq polymerase, 50 ng DNA and dH2O. The PCR thermal cycling profile included an initial incubation at 50 °C for 2 min, 15 min at 95 °C, and 40 amplification cycles (15 s at 95 °C, 1 min at 60-64 °C). Table 1 lists primers and probes. Finally, 1 μL of each LDR amplification product was run on an ABI prism 310 sequencer (Applied Biosystems, Warrington, United Kingdom), and sample genotypes were analysed using Genotyper software programmes (Applied Biosystems).
| exon 8 (c.1171G > A) | F5'-CTACAAGTGCCAGTGTGAGGAA-3' | |
| R5’_CCCACCACTCTGCTTGTAAGGCGTGAGGCCGCC-3' | ||
| Allele 1 specific probe VIC-ACACGAAGGCCTGC-NFQ | ||
| LDLR primers | Allele 2 specific probe 6-FAM-ACACGAAGACCT GC-NFQ | |
| exon 10 (c.1413G > A) | F5'- CGGCGTCTCTTCCTATGACA-3' | |
| R5'- GTCCAGTAGATGTTGCTGTGGAT-3' | ||
| Allele 1 specific probe VIC-ATCAGCAGGGAC ATC-NFQ | ||
| Allele 2 specific probe 6-FAM-TCAGCAGAGACA TC-NFQ |
DNA Sequencing: PCR product samples were sequenced with a forward primer using a Big Dye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, United States) according to the manufacturer’s instructions. Briefly, PCR sequencing cycling reactions were performed in a final total volume of 20 μL, which included 8 μL Big Dye terminator, 3.2 μL of 1 pmol diluted forward primer, 1 μL PCR product and 7.8 μL nuclease free water. The thermal profile conditions were 94 °C for 4 min, 95 °C for 15 s, 55 °C for 30 s and 60 °C for 4 min for 25 cycles. The sequencing cycling PCR products were purified using centri-sep nucleic acid gel-purified columns (Life Technology, Invitrogen). Ten microlitre of Hi Di formamide were added to 10 μL of purified PCR products. The mix was denatured in a PCR thermal cycler (Biometra, Germany) at 95 °C for 5 min. The reactions were performed in an automatic Sequencer ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, Calif.). The sequences obtained were analysed using the GenBank BLAST tool. The sequences were edited and aligned using the BioEdit Sequence Alignment. Table 1 shows the primers and probes for the LDLR genes.
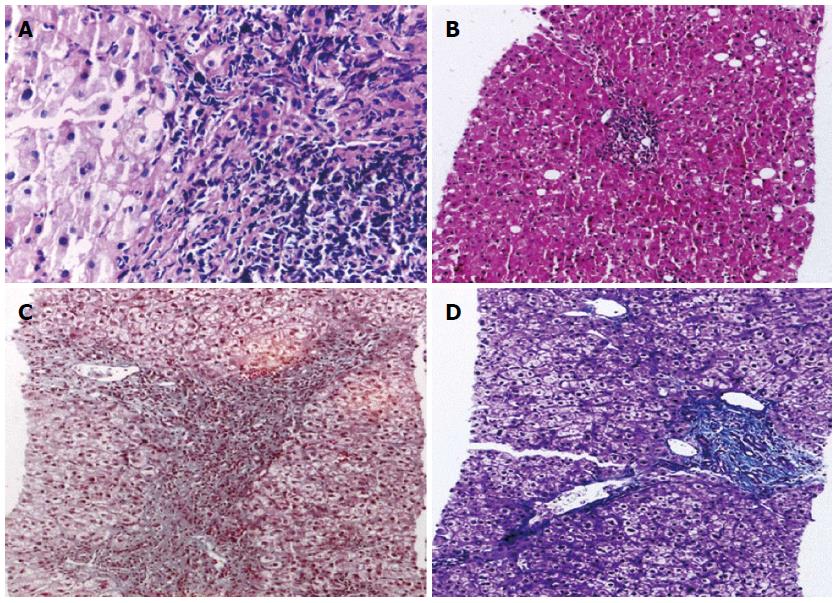
Liver biopsies were performed using 18-gauge Tru-Cut Biopsy Needles under ultrasonographic guidance with a 3.5-MHz convex probe of a GE LOGIQ® P5 ultrasound machine after 6 h of fasting under complete aseptic conditions and the use of a local anaesthesia (5 mL of 2% lidocaine). Patients’ vital signs of all patients were controlled for 2-4 h after the procedure to control probable complications. Biopsy specimens were placed in 10% formaldehyde and transferred to our pathology laboratory on the same day.
All liver core biopsies were fixed for 24 h in a 10% neutral-buffered formalin solution, processed in ascending grades of ethyl alcohol (70%, 90%, and 100%) and xylene, and placed in paraffin blocks. Three paraffin wax sections were cut at 5 microns and stained using the following reagents: (1) hematoxylin and eosin for routine histopathological examination, diagnosis and necro-inflammatory scoring; (2) Masson Trichrome to detect the extent of fibrosis. This stain imparts a blue colour to collagen against a red background of hepatocytes and other structures. It highlights the presence and distribution of reactive fibrosis as a result of liver injury. It is used for the staging of chronic liver diseases; and (3) prussian blue is a common and reliable stain for the detection of iron, which appears as blue granules in the cytoplasm.
Necro-inflammatory scoring and staging of the studied cases was performed using a modified HAI score and Metavir systems[39].
Data obtained from the study were coded and entered using SPSS (Statistical package for social science) software version 21. Parametric data are summarized using means and standard deviation, and non-parametric data are summarized as medians and percentiles for quantitative variables. Frequencies and percentages were used for qualitative variables. Comparisons between groups were performed using the χ2 and Fischer’s exact tests for qualitative variables, and Student’s t test and the non-parametric Mann-Whitney test were used to compare two groups. ANOVA and a non-parametric test (Kruskal Wallis test) were used to compare multiple groups. The odds ratio (OR) and 95%CI were calculated to estimate the strength of associations between each genotype and alleles and patients and controls. P values were considered significant when P < 0.05.
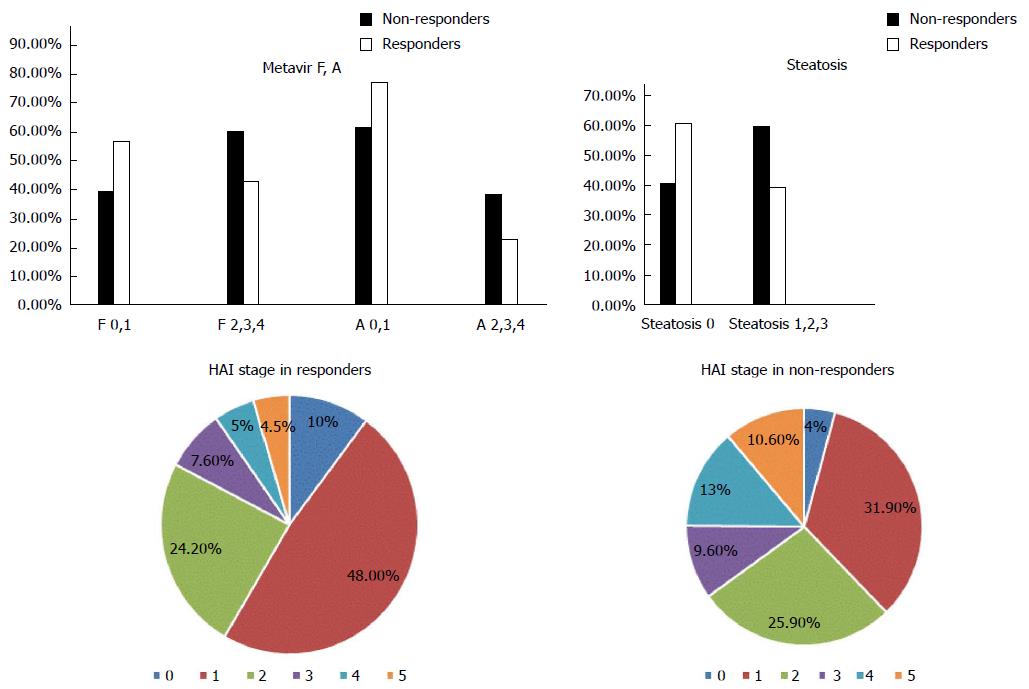
A total of 657 chronic active hepatitis C patients were used in this study. Mean patient age was 42.5 ± 10.2 years. There were 399 males and 258 females. The control population included 160 volunteers. Patients were divided into 301 non-responders and 356 responders based on their responses to therapy. Median levels of BMI, ALT, and AST were significantly higher in the non-responders vs responders. There were no statistically significant differences in levels of albumin, viral load, platelet count, or ANA, and the presence of co-morbidities or the type of interferon used between groups. Liver biopsies from patients prior to treatment revealed a significant difference between responders and non-responders. Responders exhibited fewer disturbances in liver architecture as evidenced by HAI stage and Metavir A and F. There was also a significantly greater percentage of non-responders who exhibited liver steatosis than responders (Figures 1 and 2).
The frequency of the LDLR exon 8 C: 1171 G>A in the patient group was distributed as follows: AA: 170 (25.9%), GA: 146 (22.2%), and GG: 341 (51.9%). This result was significantly different from the control group: AA: 6 (3.8%), GA: 85 (53.1%) and GG: 69 (43.1%). Responders exhibited the following frequency of LDLR exon 8C:1171 G>A polymorphism: AA: 13 (3.6%), GA: 54 (15.2%), and GG: 289 (81.2%). Non-responders exhibited the following distribution: AA: 157 (52.2%), GA: 92 (30.6%), and GG: 52 (17.2%). These differences were statistically significant (P < 0.001).
LDLR exon 10 C1413 G>A revealed no significant differences between cases and controls, and there was no difference between responders and non-responders (P = 0.354) (Table 2).
| Non responders (n = 301) | Responders (n = 356) | Control(n = 160) | P value | |
| LDR exon8 c.1171G>A | < 0.001 | |||
| AA | 157 (52.2) | 13 (3.6) | 6 (3.8) | |
| GA | 92 (30.6) | 54 (15.2) | 85 (53.1) | |
| GG | 52 (17.2) | 289 (81.2) | 69 (43.1) | |
| LDR exon10 c.1413G>A | 0.204 | |||
| AA | 48 (15.9) | 51 (14.3) | 15 (9.4) | |
| GA | 191 (63.5) | 215 (60.4) | 109 (68.1) | |
| GG | 62 (20.6) | 90 (25.3) | 36 (22.5) |
There was a statistically significant association between the A allele of LDLR exon 8 C1171 G>A gene polymorphism and the response to treatment (standard of care treatment). Carriers of the A allele exhibited a 16.4 times greater risk for non-response to treatment than carriers of the G allele (P < 0.001). There was no significant association between response to therapy and LDLR exon 10 C:1413 G>A (Table 3).
| Non responders | Responders | P value | OR (95%CI) | |
| n (%) | n (%) | |||
| LDR exon 8 c.1171G>Aallele | ||||
| A | 406 (67.4) | 80 (11.2) | < 0.001 | 16.4 (12.3-21.8) |
| G | 196 (32.6) | 632 (88.8) | 1 | |
| exon 10 c.1413 G>Aallele | ||||
| A | 287 (47.7) | 317 (44.5) | 0.253 | 1.13 (0.9-1.4) |
| G | 315 (52.3) | 395 (55.5) | 1 |
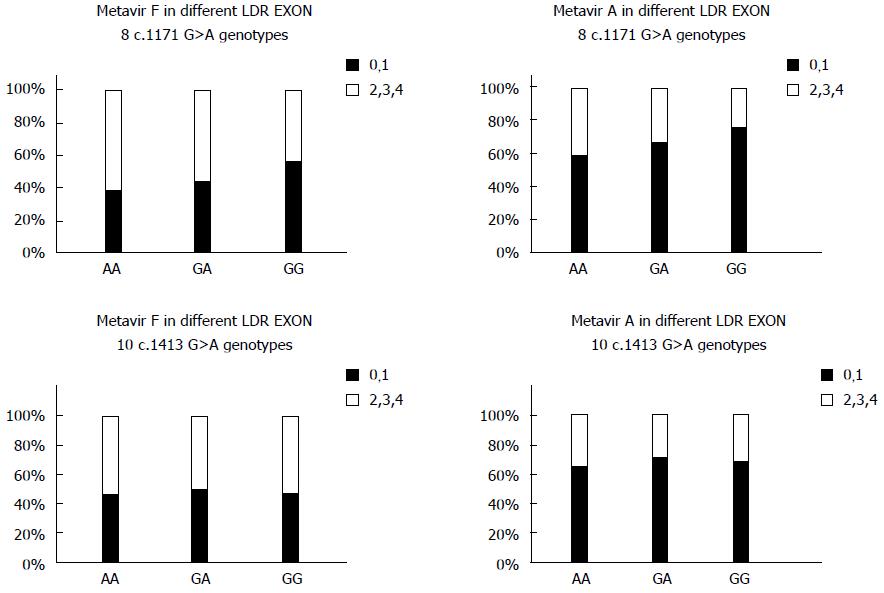
The LDLR exon 8 C1171 G>A polymorphism was significantly associated with the HAI stage of liver fibrosis. There was a statistically significant association between the AA variant of the LDLR exon 8 genotype and increased BMI. There was no significant association between the different genotypes of LDLR exon 10 polymorphisms and liver fibrosis as evidenced by HAI and Metavir scores. There was a statistically significant association between LDLR exon 8 and Metavir A and F scores and the pathological evidence of steatosis. Carriers of the AA genotype exhibited a significantly higher incidence of fibrosis and steatosis. There was no association between the various genotypes of LDLR exon 10 gene polymorphisms and Metavir A or F scores or the presence of steatosis (Figure 3 and Table 4). There was no association between LDL receptor genotypes and the type of interferon in responders and non-responders.
| LDR exon 8 c.1171 G>A | P value | LDR exon 10 c.1413 G>A | P value | |||||
| AA | GA | GG | AA | GA | GG | |||
| BMI (kg/m2) | 28.7 ± 4.7 | 28.1 ± 4.8 | 26.6 ± 4.3 | < 0.001 | 27.3 ± 4.3 | 27.5 ± 4.8 | 27.4 ± 4.4 | 0.887 |
| Steatosis | ||||||||
| 0 | 64 (37.6) | 70 (47.9) | 204 (59.8) | < 0.001 | 50 (50.5) | 208 (51.2) | 80 (52.6) | 0.938 |
| 1,2,3 | 106 (62.4) | 76 (52.1) | 137 (40.2) | 49 (49.5) | 198 (48.8) | 72 (47.4) | ||
Many studies suggested that HCV likely utilizes cell receptors to facilitate entry into cells[40]. The discrepancy in the rate of progression and clinical outcome of this disease was extensively investigated, and this step of viral entry is most incriminated in the development of chronicity[17]. The LDL receptor is a transmembrane glycoprotein that acts as a receptor for the uptake of cholesterols containing serum lipoproteins[41].
The LDL receptor is a proposed receptor for HCV entry into hepatocytes. However the exact mechanism of HCV entry is not clear. The association of the virus with a host’s lipoprotein component may facilitate this interaction[42]. The endoplasmic reticulum is the likely site of production of lipid droplets that associate with HCV after its replication and assembly[43]. Mature viruses are released from infected cells, and they are found in the low-density fraction of the serum of infected persons[44]. Hepatocytes endocytosed HCV-RNA in the LDL fraction of HCV-infected patients. Anti-LDL and anti-ApoE antibodies block viral entry into cells[17]. Several in vitro studies confirmed this hypothesis and concluded that the binding and entry of HCV particles into cultured hepatocytes was strongly associated with the level of LDLR expression[45]. HCV infection propagates LDLR expression, which further promotes lipid uptake from the blood and enriches the hepatocyte content of lipids to facilitate HCV replication[44,46].
Some studies indicate that the LDLR functions as a receptor of HCV binding and entry through interactions between the HCV/lipoprotein virion complex[47]. Other studies suggest that the LDLR is essential only for post-entry events, such as viral replication[48]. These studies note the possible role of other host receptors in HCV binding, attachment, endophagocytosis and viral replication, whether sequentially or individually. However, the viral envelope glycoproteins E1 and E2 are the main binding sites for the host receptors[49].
Direct interaction of HCVE2 envelope proteins and LDL mediates the binding and internalisation of lipoviroparticles by LDLR[32]. Atherosclerosis, obesity and familial hypercholesterolaemia are definitely related to mutations and polymorphisms in the gene encoding for LDLR[25]. This fact led investigators to probe the significance of gene polymorphisms of LDLR and the development of fibrosis, and the response to the standard of care treatment.
This study assessed gene polymorphisms at exon 8 G>A 1171 and exon 10 G>A 1413. The study was conducted on 657 patients with HCV infection and 160 healthy controls. The genotype distribution and allele frequency of exons 8 and 10 of the LDLR was studied and related to liver inflammation, the presence of steatosis, and BMI.
Our results demonstrated that the AA gene polymorphism of the 3 types of gene polymorphisms found in exon 8 of the LDLR (AA, GA, GG) was significantly different (P < 0.001) between HCV patients and the healthy control group, but the GA and GG gene polymorphisms were not significantly different. LDLR exon 10 also revealed no significance in gene polymorphisms (either AA, GA or GG), which indicates that the presence of the AA gene polymorphism of LDLR exon 8 was clearly associated with a higher susceptibility to HCV infection. Furthermore, the LDLR exon 8 AA gene polymorphism occurred more frequently in non-responders of interferon-based treatment than responders, and this difference was highly statistically significant (P < 0.001). A total of 157 patients (52.2%) of the non-responders had AA genes, but only 13 (3.6%) of the responders had AA genes.
However, no such discrepancy was detected in patients with GA or GG genes. exon 10 also exhibited no differences between gene polymorphisms between responders and non-responders.
One of the most important outcomes of the present study was the statistically significant association between the A allele of LDLR exon 8 gene polymorphism and the response to interferon, with carriers of the A allele exhibiting a 16.4 times greater risk for non-response to interferon-based treatment than patients who did not carry the A allele (P < 0.001), regardless of the type of interferon used.
The study identified a significant association between LDLR exon 8 and HAI staging and Metavir A and Metavir F, but this statistically significant difference was not present in LDLR exon 10.
Steatosis and BMI levels were significantly associated with LDLR exon 8, and both factors were higher in patients carrying the AA genotype. However, no such association was found in LDLR exon 10.
This difference may be explained by the fact that polymorphisms in the LDLR gene theoretically affect steatosis, which eventually leads to more severe liver disease and the failure of response to treatment in patients with HCV. However, the relationship between exon 8 and steatosis requires further study.
The finding that a single polymorphism in exon 8 was associated with more severe liver disease was also concluded in a study by Li et al[50] in 2006, who reported a correlation between exon 8 and fibrosis severity. In contrast to our study, Li et al[50] correlated a polymorphism of exon 10 to viral clearance and overall inflammation, which was not observed in our study. This difference may be explained by the difference in the ethnicity of the group studied and/or the type of viral genome.
The same correlation between exon 10, viral clearance and overall inflammation was also observed in a study by Hennig et al[36]. This study agreed with our study that the polymorphism in exon 8 exhibited the strongest association with disease severity. However the carriage of the G allele on exon 8 was associated with more severe fibrosis in this study, and heterozygosity at this site appeared protective against inflammation. This result directly contradicts our study in which the presence of the A allele was associated with more severe disease, and the presence of the G allele was somewhat protective.
A more recent study by Mas Marques et al[51] in 2009 demonstrated that the prevalence of an LDLR exon 8 polymorphism did not alter the grade of inflammation or stage of fibrosis. However, a careful analysis of this study reveals a low prevalence of the A allele of exon 8, which may support our results.
In conclusion, polymorphisms in the LDLR play a key role in the progression of HCV infection. The recent study of different exons identified exons 8 and 10 for further scrutiny. The results that relate polymorphisms of the alleles of these exons to the propagation of HCV infection and disease progression are somewhat contradictory. The need to clarify this dilemma is evident to improve disease outcome and treatment response in patients with HCV. This results of the present and previous studies supports the pivotal role of LDLR exon 8 in the natural course of HCV and its response to treatment. However, it is evident that further studies are needed to clarify whether other exons and alleles play a similar role.
Special thanks to Professor Nouman Elgarim (Professor of internal medicine, Cairo University) and associate professor Ahmed Abdel Moaaty for their efforts to facilitate the recruitment of patients from Beni-Suef public hospital. We thank Dr. Amr Abdelaziz (Assistant lecturer of internal medicine, Cairo University) for his efforts in sample collection from the Beni-Suef public hospital. We also thank Dr. Ismail Kassem (Assistant Professor of pathology, National Liver Institute, Cairo) for his help in the recruitment of patients from the National Liver Institute.
Hepatitis C virus (HCV) infection causes severe morbidity and mortality worldwide. Egypt has the largest epidemic of HCV worldwide. It was proposed that low-density lipoprotein receptors (LDLR) function as HCV receptors, and these receptors are implicated in the entry of HCV into hepatocytes.
Genetic polymorphisms of the LDLR may be involved in treatment response and final disease outcome. Assessment of the frequency of LDLR genotype polymorphisms and the effect of these genetic polymorphisms in Egyptian chronic HCV patients aid predictions of treatment response.
The results demonstrated that the G allele of LDL receptor exon8c.1171G>A predominated in cases and controls over the A allele. Moreover, carriers of the A allele have a 16.4 times greater risk for non-response to anti-HCV therapy. A significant association was also found between LDL receptor exon8 c.1171G>A and the degree of liver fibrosis.
These findings aid in the selection of patients who will benefit from anti-HCV therapy. The study also confirmed the important role of LDLR exon 8 in the natural course of HCV and patient response to treatment. However, further studies are needed to clarify whether other exons and alleles play a similar role.
LDLR is a cell surface receptor that plays an important role in cholesterol homeostasis. Mutations and polymorphisms in the LDLR gene are associated with familial hypercholesterolemia, obesity and atherosclerosis. The LDLR acts as a viral receptor. The LDLR gene family functions as a receptor for the minor group of common cold viruses, and these receptors may function as HCV and other virus receptors.
This report was very interesting study about HCV and LDLR polymorphism. HCV particles are known to be in complex with lipoproteins. Discussion need to be more detailed about LDLR and HCV.
P- Reviewer: Mahmood T, Ogura S S- Editor: Yu J L- Editor: A E- Editor: Liu XM
| 1. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1077] [Cited by in F6Publishing: 1066] [Article Influence: 118.4] [Reference Citation Analysis (0)] |
| 2. | Marcellin P. Hepatitis C: the clinical spectrum of the disease. J Hepatol. 1999;31 Suppl 1:9-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 169] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 3. | Lee MH, Yang HI, Yuan Y, L’Italien G, Chen CJ. Epidemiology and natural history of hepatitis C virus infection. World J Gastroenterol. 2014;20:9270-9280. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 65] [Reference Citation Analysis (1)] |
| 4. | Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10:553-562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 704] [Cited by in F6Publishing: 707] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 5. | Dumortier J, Boillot O, Scoazec JY. Natural history, treatment and prevention of hepatitis C recurrence after liver transplantation: past, present and future. World J Gastroenterol. 2014;20:11069-11079. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 20] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Lee JJ, Kim PT, Fischer S, Fung S, Gallinger S, McGilvray I, Moulton CA, Wei AC, Greig PD, Cleary SP. Impact of viral hepatitis on outcomes after liver resection for hepatocellular carcinoma: results from a north american center. Ann Surg Oncol. 2014;21:2708-2716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Lee MH, Yang HI, Lu SN, Jen CL, You SL, Wang LY, L’Italien G, Chen CJ, Yuan Y. Hepatitis C virus genotype 1b increases cumulative lifetime risk of hepatocellular carcinoma. Int J Cancer. 2014;135:1119-1126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Jahan S, Ashfaq UA, Qasim M, Khaliq S, Saleem MJ, Afzal N. Hepatitis C virus to hepatocellular carcinoma. Infect Agent Cancer. 2012;7:2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | El-Zanaty F, Way A. Egypt Demographic and Health Survey 2008. Egypt Demographic health survey. Final report, 2009. Egyptian Ministry of Health. El-Zanaty and associates and macro international: Cairo 2009; 431. [Cited in This Article: ] |
| 10. | Guerra J, Garenne M, Mohamed MK, Fontanet A. HCV burden of infection in Egypt: results from a nationwide survey. J Viral Hepat. 2012;19:560-567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 11. | Cuadros DF, Branscum AJ, Miller FD, Abu-Raddad LJ. Spatial epidemiology of hepatitis C virus infection in Egypt: analyses and implications. Hepatology. 2014;60:1150-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 12. | Reker C, Islam KM. Risk factors associated with high prevalence rates of hepatitis C infection in Egypt. Int J Infect Dis. 2014;25:104-106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Kim DD, Hutton DW, Raouf AA, Salama M, Hablas A, Seifeldin IA, Soliman AS. Cost-effectiveness model for hepatitis C screening and treatment: Implications for Egypt and other countries with high prevalence. Glob Public Health. 2015;10:296-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 14. | Benova L, Awad SF, Miller FD, Abu-Raddad LJ. Estimation of hepatitis C virus infections resulting from vertical transmission in Egypt. Hepatology. 2015;61:834-842. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Lavanchy D, McMahon B. 10 Worldwide prevalence and prevention of hepatitis C. Hepatitis C. San Diego: Academic Press 2000; 185-201 Available from: http://www.sciencedirect.com/science/article/pii/S1874532600800149. [Cited in This Article: ] |
| 16. | Abdel-Rahman M, El-Sayed M, El Raziky M, Elsharkawy A, El-Akel W, Ghoneim H, Khattab H, Esmat G. Coinfection with hepatitis C virus and schistosomiasis: fibrosis and treatment response. World J Gastroenterol. 2013;19:2691-2696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 28] [Cited by in F6Publishing: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766-12771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 702] [Cited by in F6Publishing: 688] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 18. | Monazahian M, Böhme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J Med Virol. 1999;57:223-229. [PubMed] [Cited in This Article: ] |
| 19. | Jahan S, Samreen B, Khaliq S, Ijaz B, Khan M, Siddique MH, Ahmad W, Hassan S. HCV entry receptors as potential targets for siRNA-based inhibition of HCV. Genet Vaccines Ther. 2011;9:15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Rivero-Juarez A, Camacho A, Caruz A, Neukam K, Gonzalez R, Di Lello FA, Perez-Camacho I, Mesa P, Torre-Cisneros J, Peña J. LDLr genotype modifies the impact of IL28B on HCV viral kinetics after the first weeks of treatment with PEG-IFN/RBV in HIV/HCV patients. AIDS. 2012;26:1009-1015. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 21. | Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blaas D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91:1839-1842. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 331] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 22. | Bates P, Young JA, Varmus HE. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043-1051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 282] [Cited by in F6Publishing: 286] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 23. | Go GW, Mani A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J Biol Med. 2012;85:19-28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 24. | Morales-Villegas E. PCSK9 and LDLR The Yin-Yang in the Cellular Uptake of Cholesterol. Curr Hypertens Rev. 2014;Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Li H, Zhang Y, Wei X, Peng Y, Yang P, Tan H, Chen C, Pan Q, Liang D, Wu L. Rare intracranial cholesterol deposition and a homozygous mutation of LDLR in a familial hypercholesterolemia patient. Gene. 2015;569:313-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | De Castro-Orós I, Pérez-López J, Mateo-Gallego R, Rebollar S, Ledesma M, León M, Cofán M, Casasnovas JA, Ros E, Rodríguez-Rey JC. A genetic variant in the LDLR promoter is responsible for part of the LDL-cholesterol variability in primary hypercholesterolemia. BMC Med Genomics. 2014;7:17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | Khan SP, Ghani R, Ahmed KZ, Yaqoob Z. Two novel mutations in exon 3 and 4 of low density lipoprotein (LDL) receptor gene in patients with heterozygous familial hypercholesterolemia. J Coll Physicians Surg Pak. 2011;21:403-406. [PubMed] [Cited in This Article: ] |
| 28. | Zee RY, Schrader AP, Robinson BG, Griffiths LR, Morris BJ. Association of HincII RFLP of low density lipoprotein receptor gene with obesity in essential hypertensives. Clin Genet. 1995;47:118-121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 29. | Sarzani R, Salvi F, Bordicchia M, Guerra F, Battistoni I, Pagliariccio G, Carbonari L, Dessì-Fulgheri P, Rappelli A. Carotid artery atherosclerosis in hypertensive patients with a functional LDL receptor-related protein 6 gene variant. Nutr Metab Cardiovasc Dis. 2011;21:150-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Ding Z, Liu S, Wang X, Khaidakov M, Fan Y, Deng X, Xiang D, Mehta JL. Lectin-like oxidized low-density lipoprotein receptor-1 regulates autophagy and Toll-like receptor 4 in the brain of hypertensive mice. J Hypertens. 2015;33:525-533; discussion 533. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Salazar LA, Hirata MH, Giannini SD, Forti N, Diament J, Lima TM, Hirata RD. Seven DNA polymorphisms at the candidate genes of atherosclerosis in Brazilian women with angiographically documented coronary artery disease. Clin Chim Acta. 2000;300:139-149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Wunschmann S, Muller HM, Stipp CS, Hemler ME, Stapleton JT. In vitro interaction between hepatitis C virus (HCV) envelope glycoprotein E2 and serum lipoproteins (LPs) results in enhanced cellular binding of both HCV E2 and LPs. J Infect Dis. 2006;194:1058-1067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Zhao LJ, Zhao P, Chen QL, Ren H, Pan W, Qi ZT. Mitogen-activated protein kinase signalling pathways triggered by the hepatitis C virus envelope protein E2: implications for the prevention of infection. Cell Prolif. 2007;40:508-521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 34. | Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411-419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 214] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 35. | Carrière M, Rosenberg AR, Conti F, Chouzenoux S, Terris B, Sogni P, Soubrane O, Calmus Y, Podevin P. Low density lipoprotein receptor transcripts correlates with liver hepatitis C virus RNA in patients with alcohol consumption. J Viral Hepat. 2006;13:633-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 36. | Hennig BJ, Hellier S, Frodsham AJ, Zhang L, Klenerman P, Knapp S, Wright M, Thomas HC, Thursz M, Hill AV. Association of low-density lipoprotein receptor polymorphisms and outcome of hepatitis C infection. Genes Immun. 2002;3:359-367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 56] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 37. | Matas M, Picornell A, Cifuentes C, Payeras A, Homar F, González-Candelas F, López-Labrador FX, Moya A, Ramon C, Castro JA. Relating the outcome of HCV infection and different host SNP polymorphisms in a Majorcan population coinfected with HCV-HIV and treated with pegIFN-RBV. Int Microbiol. 2014;17:11-20. [PubMed] [Cited in This Article: ] |
| 38. | Soriano V, Poveda E, Vispo E, Labarga P, Rallón N, Barreiro P. Pharmacogenetics of hepatitis C. J Antimicrob Chemother. 2012;67:523-529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Lefkowitch JH. Liver biopsy assessment in chronic hepatitis. Arch Med Res. 2007;38:634-643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 40. | Helle F, Dubuisson J. Hepatitis C virus entry into host cells. Cell Mol Life Sci. 2008;65:100-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 41. | Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431-438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 804] [Cited by in F6Publishing: 847] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 42. | Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120-2129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 347] [Cited by in F6Publishing: 349] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 43. | Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089-1097. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 937] [Cited by in F6Publishing: 948] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 44. | Syed GH, Tang H, Khan M, Hassanein T, Liu J, Siddiqui A. Hepatitis C virus stimulates low-density lipoprotein receptor expression to facilitate viral propagation. J Virol. 2014;88:2519-2529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 45. | Martin C, Nielsen SU, Ibrahim S, Bassendine MF, Toms GL. Binding of liver derived, low density hepatitis C virus to human hepatoma cells. J Med Virol. 2008;80:816-823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 46. | Caruz A, Neukam K, Rivero-Juárez A, Herrero R, Real LM, Camacho A, Barreiro P, Labarga P, Rivero A, Pineda JA. Association of low-density lipoprotein receptor genotypes with hepatitis C viral load. Genes Immun. 2014;15:16-24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 47. | Owen DM, Huang H, Ye J, Gale M. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 48. | Albecka A, Belouzard S, Op de Beeck A, Descamps V, Goueslain L, Bertrand-Michel J, Tercé F, Duverlie G, Rouillé Y, Dubuisson J. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology. 2012;55:998-1007. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 49. | Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017-5025. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 888] [Cited by in F6Publishing: 868] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 50. | Li H, Liu Z, Han Q, Li Y, Chen J. Association of genetic polymorphism of low-density lipoprotein receptor with chronic viral hepatitis C infection in Han Chinese. J Med Virol. 2006;78:1289-1295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 51. | Mas Marques A, Mueller T, Welke J, Taube S, Sarrazin C, Wiese M, Halangk J, Witt H, Ahlenstiel G, Spengler U. Low-density lipoprotein receptor variants are associated with spontaneous and treatment-induced recovery from hepatitis C virus infection. Infect Genet Evol. 2009;9:847-852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |