Published online Sep 15, 2004. doi: 10.3748/wjg.v10.i18.2619
Revised: August 24, 2003
Accepted: October 7, 2003
Published online: September 15, 2004
AIM: To construct a phage display library of human single-chain variable fragment (scFv) antibodies associated with esophageal cancer and to preliminarily screen a scFv antibody against esophageal cancer.
METHODS: Total RNA extracted from metastatic lymph nodes of esophageal cancer patients was used to construct a scFv gene library. Rescued by M13K07 helper phage, the scFv phage display library was constructed. esophageal cancer cell line Eca109 and normal human esophageal epithelial cell line (NHEEC) were used for panning and subtractive panning of the scFv phage display library to obtain positive phage clones. Soluble scFv was expressed in E. coli HB2151 which was transfected with the positive phage clone, then purified by affinity chromatography. Relative molecular mass of soluble scFv was estimated by Western blotting, its bioactivity was detected by cell ELISA assay. Sequence of scFv was determined using the method of dideoxynucleotide sequencing.
RESULTS: The size of scFv gene library was approximately 9 × 106 clones. After four rounds of panning with Eca109 and three rounds of subtractive panning with NHEEC cells, 25 positive phage clones were obtained. Soluble scFv was found to have a molecular mass of 31 ku and was able to bind to Eca109 cells, but not to HeLa and NHEEC cells. Variable heavy (VH) gene from one of the positive clones was shown to be derived from the γ chain subgroup IV of immunoglobulin, and variable light (VL) gene from the κ chain subgroup I of immunoglobulin.
CONCLUSION: A human scFv phage display library can be constructed from the metastatic lymph nodes of esophageal cancer patients. A whole human scFv against esophageal cancer shows some bioactivity.
- Citation: Xu MY, Xu XH, Chen GZ, Deng XL, Li J, Yu XJ, Chen MZ. Production of a human single-chain variable fragment antibody against esophageal carcinoma. World J Gastroenterol 2004; 10(18): 2619-2623
- URL: https://www.wjgnet.com/1007-9327/full/v10/i18/2619.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i18.2619
Esophageal cancer is one of the most common malignancies in China with a relatively high mortality rate. In recent years, antibodymediated tumor immunoscintigraphy and immunotherapy have been used in the diagnostic and therapeutic approaches of cancers[1,2]. However, most antibodies are of murine origin, and repeated administration can induce human anti-mouse antibodies (HAMA). In addition, intact antibody is too large to penetrate into tumor masses, its application is limited. To overcome such deficiencies, many kinds of humanized antibodies including human-murine chimeric antibody and small molecular antibodies have been developed, but they are still of murine origin.
Recently, the emergence of genetically engineered antibodies and phage display libraries of human antibody fragments from immune or naïve donors has enabled the production of human antibody fragment targeting cancers[3]. In the present study, phage antibody library techniques were used to construct a human phage single-chain Fv antibody library from metastatic lymph nodes of esophageal cancer patients. To obtain a single chain Fv AD09, panning and subtractive panning were performed with human esophageal cancer cell line (Eca109) and normal human esophageal epithelial cell line (NHEEC) respectively. Soluble AD09 was expressed in E. coli HB2151 and purified by affinity chromatography using anti-E tag antibody, its bioactivity was then detected by cell ELISA assay.
Human esophageal carcinoma cell line Eca109 (Cytology Institute of Chinese Medical Academy, Beijing) and HeLa cell line (Shanghai Cytology Institute, China) were cultured at 37 °C in RPMI1640 medium supplemented with 100 mL/L fetal calf serum (Hyclone, USA) in a humidified atmosphere of 50 mL/L CO2. Normal human esophageal epithelial cell (NHEEC) line was a primary cell line from a 20-wk conception fetus cultured in RPMI1640 with 200 mL/L fetal calf serum.
Primer sequences were created as previously described[4] with some modifications in PCR assembly part (Table 1B). We designed complementary coding sequences for a peptide linker at the 5’-end of JH forward primers and the 3’-end of human V kappa (or lambda) back primers to optimize the diversity and efficiency of ligation. The primers were synthesized by Sunbiotech Company (Beijing, China) and the sequences are shown in Table 1. Sequences were given using the IUPAC nomenclature of mixed base (R = A or T, K = G or T, Y = C or T, S = G or C, H = A or C or T, N = A or C or G or T).
| A. Primary PCRs | |
| Human VH back primers (sense) | |
| HuVHlaBACK | 5’-CAGGTGCAGCTGGTGCAGTCTGG-3’ |
| HuVH2aBACK | 5’-CAGGTCAACTTAAGGGAGTCTGG-3’ |
| HuVH3aBACK | 5’-GAGGTGCAGCTGGTGGAGTCTGG-3’ |
| HuVH4aBACk | 5’-CAGGTGCAGCTGCAGGAGTCGGG-3’ |
| HuVH5aBACK | 5’-GAGGTGCAGCTGTTGCAGTCTGC-3’ |
| HuVH6aBACK | 5’-CAGGTACAGCTGCAGCAGTCAGG-3’ |
| JH forward primers (anti-sense) | |
| HuJHl-2FOR | 5’-TGAGGAGACGGTGACCAGGGTGCC-3’ |
| HuJH3FOR | 5’-TGAAGAGACGGTGACCATTGTCCC-3’ |
| HuJH4-5FOR | 5’-TGAGGAGACGGTGACCAGGGTTCC-3’ |
| HuJH6FOR | 5’-TGAGGAGACGGTGACCGTGGTCCC-3’ |
| Human V kappa back primers (sense) | |
| HuV κ laBack | 5’-GACATCCAGATGACCCAGTCTCC-3’ |
| HuV κ 2aBack | 5’-GATGTTGTGATGACTCAGTCTCC-3’ |
| HuV κ 3aBack | 5’-GAAATTGTGTTGACGCAGTCTCC-3’ |
| HuV κ 4aBack | 5’-GACATCGTGATGACCCAGTCTCC-3’ |
| HuV κ 5aBack | 5’-GAAACGACACTCACGCAGTCTCC-3’ |
| HuV κ 6aBack | 5’-GAAATTGTGCTGACTCAGTCTCC-3’ |
| Human J kappa forward primer (anti-sense) | |
| HuJ κ 1FOR | 5’-ACGTTTGATTTCCACCTTGGTCCC-3’ |
| HuJ κ 2FOR | 5’-ACGTTTGATCTCCAGCTTGGTCCC-3’ |
| HuJ κ 3FOR | 5’-ACGTTTGATATCCACTTTGGTCCC-3’ |
| HuJ κ 4FOR | 5’-ACGTTTGATCTCCACCTTGGTCCC-3’ |
| HuJ κ 5FOR | 5’-ACGTTTAATCTCCAGTCGTGTCCC-3’ |
| Human V lambda back primers (sense) | |
| HuV λ 1BACK | 5’-CAGTCTGTGTTGACGCAGCCGCC-3’ |
| HuV λ 2BACK | 5’-CAGTCTGCCCTGACTCAGCCTGC-3’ |
| HuV λ 3aBACK | 5’-TCCTATGTGCTGACTCAGCCACC-3’ |
| HuV λ 3bBACK | 5’-TCTTCTGAGCTGACTCAGGACCC-3’ |
| HuV λ 4BACK | 5’-CACGTTATACTGACTCAACCGCC-3’ |
| HuV λ 5BACK | 5’-CAGGCTGTGCTCACTCAGCCGTC-3’ |
| HuV λ 6BACK | 5’-AATTTTATGCTGACTCAGCCCCA-3’ |
| Human J lambda forward primers (anti-sense) | |
| HuJ λ 1FOR | 5’-ACCTAGGACGGTGACCTTGGTCCC-3’ |
| HuJ λ 2-3FOR | 5’-ACCTAGGACGGTCAGCTTGGTCCC-3’ |
| HuJ λ 4-5FOR | 5’-ACCTAAAACGGTGAGCTGGGTCCC-3’ |
| B. PCR assembly | |
| Hu JH-Linker primers | |
| HuJH1-2Linker | 5’-AGAGCCACCTCCGCCTGAACCGCCTCCACCTGAGGAGACGGTGACCAGGGTGCC-3’ |
| HuJH3Linker | 5’-AGAGCCACCTCCGCCTGAACCGCCTCCACCTGAAGAGACGGTGACCATTGTCCC-3’ |
| HuJH4-5Linker | 5’-AGAGCCACCTCCGCCTGAACCGCCTCCACCTGAGGAGACGGTGACCAGGGTTCC-3’ |
| HuJH6Linker | 5’-AGAGCCACCTCCGCCTGAACCGCCTCCACCTGAGGAGACGGTGACCGTGGTCCC-3’ |
| Linker-Hu V κ primers | |
| Linker-HuV κ 2-3-6 BACK | 5’-GTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGGAWRTTGTGHTGACKCAGTCTCC-3’ |
| Linker-HuV κ 1-4 BACK | 5’-gTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGGACATCSWGATGACCCAGTCTCC-3’ |
| Linker-HuV κ 5 BACK | 5’-GTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGGAAACGACACTCACGCAGTCTCC-3’ |
| Linker-Hu V λ primers | |
| Linker-HuV λ 1-2BACK | 5’-GTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGCAGTCTGYSYTGACKCAGCCKSC-3’ |
| Linker-HuV λ 3BACK | 5’-GTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGTCYTMTGWGCTGACTCAGSMMCC-3’ |
| Linker-HuV λ 4-5BACK | 5’-GTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGCASGYTRTRCTSACTCARCCGYC-3’ |
| Linker-HuV λ 6BACK | 5’-GTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGAATTTTATGCTGACTCAGCCCCA-3’ |
| C. Reamplification with primers containing restriction sites | |
| Human VH back (Sfi) primers (sense) | |
| HuVH1aBACKSfi | 5’-GTCCTCGCAACTGCGGCCCAGCCGGCCATGGCCCAGGTGCAGCTGGTGCAGTCTGG-3’ |
| HuVH2aBACKSfi | 5’-GTCCTCGCAACTGCGGCCCAGCCGGCCATGGCCCAGGTCAACTTAAGGGAGTCTGG-3’ |
| HuVH3aBACKSfi | 5’-GTCCTCGCAACTGCGGCCCAGCCGGCCATGGCCGAGGTGCAGCTGGTGGAGTCTGG-3’ |
| HuVH4aBACKSfi | 5’-GTCCTCGCAACTGCGGCCCAGCCGGCCATGGCCCAGGTGCAGCTGCAGGAGTCGGG-3’ |
| HuVH5aBACKSfi | 5’-GTCCTCGCAACTGCGGCCCAGCCGGCCATGGCCCAGGTGCAGCTGTTGCAGTCTGC-3’ |
| HuVH6aBACKSfi | 5’-GTCCTCGCAACTGCGGCCCAGCCGGCCATGGCCCAGGTACAGCTGCAGCAGTCAGG-3’ |
| Human J kappa forward (Not) primers (anti-sense) | |
| HuJ κ 1FORNot | 5’-GAGTCATTCTCGACTTGCGGCCGCACGTTTGATTTCCACCTTGGTCCC-3’ |
| HuJ κ 2FORNot | 5’-GAGTCATTCTCGACTTGCGGCCGCACGTTTGATCTCCAGCTTGGTCCC-3’ |
| HuJ κ 3FORNot | 5’-GAGTCATTCTCGACTTGCGGCCGCACGTTTGATATCCACTTTGGTCCC-3’ |
| HuJ κ 4FORNot | 5’-GAGTCATTCTCGACTTGCGGCCGCACGTTTGATCTCCACCTTGGTCCC-3’ |
| HuJ κ 5FORNot | 5’-GAGTCATTCTCGACTTGCGGCCGCACGTTTAATCTCCAGTCGTGTCCC-3’ |
| Human J lambda forward (Not) primers (anti-sense) | |
| HuJ λ 1FORNot | 5’-GAGTCATTCTCGACTTGCGGCCGCACCTAGGACGGTGACCTTGGTCCC-3’ |
| HuJ λ 2-3FORNot | 5’-GAGTCATTCTCGACTTGCGGCCGCACCTAGGACGGTCAGCTTGGTCCC-3’ |
| HuJ λ 4-5FORNot | 5’-GAGTCATTCTCGACTTGCGGCCGCACCTAAAACGGTGAGCTGGGTCCC-3’ |
Metastatic lymph nodes of 5 esophageal cancer patients were collected for total RNA extraction (TRizol, Gibco BRL, UK). First-strand cDNA synthesis was performed in the presence of 40 U RNase inhibitor, 200 U Superscript II transcriptase (Gibco BRL, UK). The sample was finally treated with 2 U RNase H for 30 min at 37 °C and stored at -20 °C until use.
IgG-specific variable heavy (VH) and light (VL) chain gene fragments were amplified using Pyrobest PCR system (TarkaRa Biotechnology, Dalian, China) for 30 cycles (at 94 °C for 30 s, at 55 °C for 30 s and at 72 °C for 1 min), with each forward oligonucleotide primer and one of the back primers (Table 1A). The fragments were isolated from a 15 g/L agarose gel with the QIAex kit (QIAgen, Germany). Then fragments were used as templates for PCR amplification to extend a linker, VH fragments used human JH-linker primers and human VH back primers, VL fragments used linker-human Vκ primers (or linker-human Vλ primers) and human Jκ forward primers (or human Jλ forward primers), respectively.
The amplified VH-linker and VL-linker PCR products were combined in a SOE-PCR reaction mixture. First, approximately 100 ng each of VH-linker and VL-linker was assembled by PCR without primers in which the short regions of complementarities built into the ends of primers drove hybridization of various fragments. An initial denaturation step (at 94 °C for 5 min) was followed by five cycles (at 94 °C for 1 min, at 60 °C for 1 min and at 72 °C for 1.5 min) in the absence of primers. After the outer primers containing restriction sites (Table 1C) were added, 30 cycles (at 94 °C for 30 s, at 60 °C for 1 min and at 72 °C for 1.5 min) were performed. These were gel-purified, digested with SfiI and NotI (TarkaRa Biotechnology, Dalian), cloned into the vector pCANTAB 5E (Amersham Pharmacia Biotech, Sweden) and transformed into electrocompetent E. coli TG1 (Amersham Pharmacia Biotech, Sweden). After electroporation, cells were plated on SOBAG medium (containing 20 g/L glucose and 100 mg/mL ampicillin) in 20 dishes and incubated overnight at 30 °C. The clones were scraped off the plates in 50 mL 2 × YT medium with 100 mL/L glycerol and subsequently stored at -70 °C. Plasmid DNA was prepared from 10 randomly selected clones using Qiagen plasmid minikit (Qiagen, Germany). PCR and a SfiI/Not I double digestion reaction were performed to identify the positive insert clones.
Ampicillin-resistant colonies were scraped into 2 × YT medium and superinfected by M13K07 (Amersham Pharmacia Biotech, Sweden) helper phage. After an overnight induction in 2 × YTAK medium without glucose, the phage preparation was precipitated in 40 g/L PEG/0.5 mol/L NaCl and resuspended in 10 g/L PBS.
To screen the positive phage clones, live Eca109 cell line as the antigen was used for panning. Live NHEEC was used for subtractive panning. The panning procedure was carried out essentially as described previously[5]. After four rounds of panning and three rounds of subtractive panning, the unabsorbed phages were amplified.
To detect the scFv-phage recombinant antibody, cell ELISA was performed. Eca109 cells (5 × 104) as antigens were grown in 96-well plates at 37 °C for 24 h, then fixed with 25 g/L glutaradehyde and blocked with 10 g/L BSA. This was followed by incubation with scFv-phage at 37 °C for 2 h. After washed three times with PBS, HRP/anti-M13 monoclonal conjugate (1:5000) (Amersham Pharmacia Biotech, Sweden) was added into wells with scFv-phage and they were incubated for 1 h at 37 °C. After washed again as above, 1 × ABTS substrate solution was added, and incubated in darkness for 30 min and the reactions were read at 405 nm. PBS was used as negative control. The absorbance reading for the positive was 0.2 or above at least three times higher than that for the negative control.
To produce soluble scFvs, strongly positive recombinant phage clones were used to infect log-phage E. coli.HB2151 (Amersham Pharmacia Biotech, Sweden). Expression of soluble scFv was induced by adding isopropyl β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mmol/L and the cultures grown overnight at 30 °C. The induced culture was centrifuged at 1500 r/min for 20 min. Cell pellets were resuspended in 2% of culture volume ice-cold 1 × TES. Subsequently, 3% of culture volume ice-cold 0.2 × TES was added, the mixture was incubated on ice for 30 min to induce a mild osmotic shock. The contents were centrifuged at 12000 g for 10 min. The supernatant, containing the soluble antibodies from the periplasm, was transferred to the clean tubes and stored at -20 °C.
Soluble scFvs from periplasm were purified by affinity chromatography. Anti-E tag antibody (Amersham Pharmacia Biotech, Sweden) was covalently coupled to a protein G column (Amersham Pharmacia Biotech, Sweden) and soluble scFvs were selected by binding to anti-E tag antibody. After washed with 20 mmol/L phosphate buffer, pH7.0, + 0.5 g/L NaN3, scFvs were eluted from the column with 0.1 mol/L glycine-HCl, pH3.0, and neutralized immediately with 1 mol/L Tris/HCl, pH8.2, + 0.5 g/L NaN3. Column fractions were assayed and positive fractions were pooled and stored at -70 °C.The expressed soluble scFv proteins were analyzed by 120 g/L sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and confirmed by Western blotting. Purity and concentration of proteins were determined with Bradford assay.
To detect the activity of soluble scFv, HRP/anti-E tag antibody (Amersham Pharmacia Biotech, Sweden) was used. Eca109 cells, HeLa cells and NHEEC cells (5 × 104) were used as antigens. The cell ELISA assay procedure was performed as described above.
Plasmid DNA was prepared from recombinant clones using the Qiagen plasmid minikit (Qiagen, Germany). Nucleic acid sequencing was carried out on the ABI PRISM 377 DNA sequencer by the method of dideoxynucleotide sequencing. DNA and deduced amino acid sequence were compared with NCBI database.
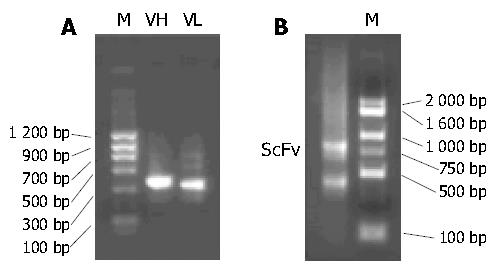
The presence of VH and VL gene fragments obtained by RT-PCR was confirmed by electrophoresis, with their sizes being approximately 350 bp (Figure 1A). The scFv genewas assembled successfully. Its size was about 750 bp (Figure 1B).
After the scFv gene repertoires were transformed into E. coli TG1 cells, approximately 9 × 106 ampicillin-resistant clones grew. PCR and SfiI/NotI double digestion reactions showed the positive insert ratio was about 95% (19/20). Rescued by M13K07 helper phage, the recombinant phage antibody library (about 9 × 1011 cfu/mL) was constructed.
Four rounds of panning with Eca109 cells resulted in a 130-fold enrichment of tumor cell binding scFv-phage. After three rounds of subtractive panning with NHEEC cells, individual phage-displayed scFv fragments were tested for reactivity with Eca109 cells by cell ELISA. Of the 95 clones screened, 25 were positive. The highest A405nm value was found in AD09 clone.
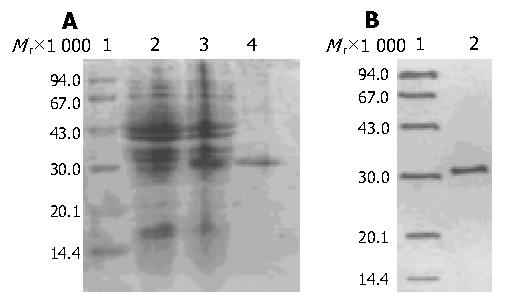
The soluble scFv was stably expressed in E. coli HB2151 transfected with AD09 positive phage clone. In pCANTAB 5E, the pel B signal peptide upstream from the scFv directed the expression to periplasmic compartment. The periplasmic extract of AD09 was run through anti-E tag antibody affinity chromatography column and soluble AD09 was eluted from the column as a single peak (data not shown). The expressed and purified AD09 was loaded on 120 g/L SDS-PAGE and analyzed by Western blot. This protein migrated with a molecular mass approximately 31 ku (Figure 2). The overall yield of soluble AD09 in E. coli flask culture was more than 0.55 mg/L. The purity was about 90%.
The immunoreactivity of purified soluble AD09 was determined by ELISA. The result revealed that AD09 was highly specific and could bind to Eca109 cells, but not to HeLa and NHEE cells (Table 2).
Sequencing of six randomly selected antibodies from the positive clones was performed with an ABI PRISM 377 DNA sequencer using pCANTAB5 sequencing primer set (Amersham Pharmacia Biotech, Sweden). The sequence f AD09 clone is shown in Figure 3. Compared using BLAST, both VH and VL had sequence similarities to the variable fragments of some known human antibodies. Alignment with the VH and VL sequences of Ig, blast analysis of immunoglobulin sequences showed that VH was the γ chain subgroup IV of human immunoglobulin and VL was the κ chain subgroup I of human immunoglobulin.
The discovery of hybridoma technology by Kohler and Milstein in 1975[6] has heralded a new era in antibody research and clinical development. However, until recently, there were few antibody-based products suitable for clinical trial. This delay can be largely explained by the fact that murine antibodies could trigger a human anti-mouse antibody response[7-9]. Human monoclonal antibodies seldom triggered a harmful immune response and have been used in cancer immunotherapy[10]. However, there are many difficulties in making human monoclonal antibodies by hybridoma technology. Phage display antibody library technique provides a powerful tool to produce human antibody. In this study, we selected a human single chain antibody against esophageal cancer from a phage displayed antibody library. It confirmed that this technique was feasible.
Since scFv could penetrate faster and deeper in solid tumors, scFv format of monoclonal antibody was selected. ScFv is a small antigen-binding antibody fragment consisting of VH and VL joined together by a flexible peptide linker. The main advantage of scFv over intact whole IgG or Fabs was its small size (one sixth of intact IgG), making it penetrate a solid tumor mass more rapidly and evenly[11,12].In addition, the lack of constant regions decreased retention by Fc receptors found in most tissues and organs, which further reduced the side effects[13,14].These characteristics rendered scFv an ideal vector for delivery of agents such as radionuclide, enzyme, drugs or toxin in vivo[14,15].
Normally, peripheral blood lymphocytes were the main source for the construction of antibody libraries[16]. However, it was inconvenient to isolate at least 200 mL of peripheral blood to obtain 107 B-lymphocytes for constructing a large antibody library[3]. There are thousands of B cells in metastatic peritumor lymph nodes which may be preimmunized by tumor-associated antigens in esophageal cancer patients. The preimmunized B cells are sufficient to construct a library, and can be directly used for screening recombinant antibodies, since the heavy and light chain genes have been rearranged and ligated to specific targets of tumor antigens. Therefore, it has become a better source of B cells for human antibody library construction[17].
Recombinant antibodies in phage antibody library could be best captured with purified tumor antigens[18]. Unfortunately, esophageal cancer associated specific antigens have not yet been identified. Human esophageal cancer cell line, Eca109 could express human esophageal cancer-associated specific antigens[19]. In addition, live cells could facilitate the identification of antibodies better than fixed cells[5,20] since the antibodies bound to native rather than denatured antigens. Moreover, using live cancer cells to screen phage antibody library was a feasible method[20,21]. So we used live Eca109 as antigens to screen the phage antibody library, and live NHEEC cell line was used for subtractive panning to obviate the cross-reaction with normal human esophageal epithelial cells. This was done to facilitate subsequent expression cloning of corresponding antigens, as well as to enhance the therapeutic potential of the antibodies obtained.
Generation of large repertoires of scFv genes is a crucial step during phage antibody display. To construct a large scFv gene repertoire, a number of methods were used. (1) Multiple primers covering most of the immunoglobulin heavy and light chain variable genes were used. (2) To optimize the diversity and efficiency of ligation, linker sequences were designed in PCR assembly primers. These made the linker easily synthesized and increased the diversity of scFv genes. (3) Electroporation transformation was used to obtain an efficiency of 109 cfu/ug for pUC18 and 3 × 107 cfu/ug for ligation products. (4) The quality of mRNA appeared essential in the PCR amplification step and in subsequent construction of the library. To preserve intact mRNA, DEPC and RNase inhibitor were used during total RNA extraction and cDNA synthesis.
To further identify the bioactivity of scFv, soluble scFv protein was expressed in E. coli HB2151 induced by IPTG. SDS-PAGE and Western blot showed that the molecular mass was about 31 ku, which was consistent with the theoretically predicted product. The soluble expression level of scFv in E. coli HB2151 was still low, but it was sufficient for bioactivity detection. Cell ELISA assays showed that the soluble scFv had esophageal cancer associated antigen-binding activity. Whether this scFv shows affinity and specificity for tissue in vivo remains to be determined. Finally, DNA sequencing determined that scFv had common characteristics shared by other known scFvs[3].
Our study demonstrated that specific human antibodies against tumor-associated antigens could be selected from a phage library constructed from the metastatic lymph nodes of esophageal cancer patients. The approach did not depend on immunization procedures. Since the antibody is entirely of human origin, it is expected to be much less immunogenic than murine antibodies and more efficient in targeting tumor cell surface. It may also be used as research reagents or a starting point for the development of therapeutic antibodies.
Edited by Zhu LH and Wang XL Proofread by Xu FM
| 1. | Souriau C, Hudson PJ. Recombinant antibodies for cancer diagnosis and therapy. Expert Opin Biol Ther. 2003;3:305-318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 2. | Trikha M, Yan L, Nakada MT. Monoclonal antibodies as therapeutics in oncology. Curr Opin Biotechnol. 2002;13:609-614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 3. | He J, Zhou G, Liu KD, Qin XY. Construction and preliminary screening of a human phage single-chain antibody library associated with gastric cancer. J Surg Res. 2002;102:150-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581-597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1184] [Cited by in F6Publishing: 1145] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 5. | Cai X, Garen A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: selection of specific antibodies from single-chain Fv fusion phage libraries. Proc Natl Acad Sci USA. 1995;92:6537-6541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 151] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 6. | Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. 1975. Biotechnology. 1992;24:524-526. [PubMed] [Cited in This Article: ] |
| 7. | Isaacs JD. The antiglobulin response to therapeutic antibodies. Semin Immunol. 1990;2:449-456. [PubMed] [Cited in This Article: ] |
| 8. | Berkower I. The promise and pitfalls of monoclonal antibody therapeutics. Curr Opin Biotechnol. 1996;7:622-628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Frödin JE, Lefvert AK, Mellstedt H. The clinical significance of HAMA in patients treated with mouse monoclonal antibodies. Cell Biophys. 1992;21:153-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 10. | Nilsson F, Tarli L, Viti F, Neri D. The use of phage display for the development of tumour targeting agents. Adv Drug Deliv Rev. 2000;43:165-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 77] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Verhaar MJ, Chester KA, Keep PA, Robson L, Pedley RB, Boden JA, Hawkins RE, Begent RH. A single chain Fv derived from a filamentous phage library has distinct tumor targeting advantages over one derived from a hybridoma. Int J Cancer. 1995;61:497-501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Mao S, Gao C, Lo CH, Wirsching P, Wong CH, Janda KD. Phage-display library selection of high-affinity human single-chain antibodies to tumor-associated carbohydrate antigens sialyl Lewisx and Lewisx. Proc Natl Acad Sci USA. 1999;96:6953-6958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 77] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 13. | Yamaguchi A, Ding K, Maehara M, Goi T, Nakagawara G. Expression of nm23-H1 gene and Sialyl Lewis X antigen in breast cancer. Oncology. 1998;55:357-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Clark M. Antibody humanization: a case of the 'Emperor's new clothes'. Immunol Today. 2000;21:397-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 195] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 15. | Kim DJ, Chung JH, Ryu YS, Rhim JH, Kim CW, Suh Y, Chung HK. Production and characterisation of a recombinant scFv reactive with human gastrointestinal carcinomas. Br J Cancer. 2002;87:405-413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 16. | Hoogenboom HR, Winter G. By-passing immunisation. Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J Mol Biol. 1992;227:381-388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 305] [Cited by in F6Publishing: 321] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 17. | Yip YL, Hawkins NJ, Clark MA, Ward RL. Evaluation of different lymphoid tissue sources for the construction of human immunoglobulin gene libraries. Immunotechnology. 1997;3:195-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Poul MA, Becerril B, Nielsen UB, Morisson P, Marks JD. Selection of tumor-specific internalizing human antibodies from phage libraries. J Mol Biol. 2000;301:1149-1161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 145] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Hao MW, Liang YR, Liu YF, Liu L, Wu MY, Yang HX. Transcription factor EGR-1 inhibits growth of hepatocellular carcinoma and esophageal carcinoma cell lines. World J Gastroenterol. 2002;8:203-207. [PubMed] [Cited in This Article: ] |
| 20. | Ridgway JB, Ng E, Kern JA, Lee J, Brush J, Goddard A, Carter P. Identification of a human anti-CD55 single-chain Fv by subtractive panning of a phage library using tumor and nontumor cell lines. Cancer Res. 1999;59:2718-2723. [PubMed] [Cited in This Article: ] |
| 21. | Kupsch JM, Tidman NH, Kang NV, Truman H, Hamilton S, Patel N, Newton Bishop JA, Leigh IM, Crowe JS. Isolation of human tumor-specific antibodies by selection of an antibody phage library on melanoma cells. Clin Cancer Res. 1999;5:925-931. [PubMed] [Cited in This Article: ] |