Published online Mar 28, 2009. doi: 10.3748/wjg.15.1480
Revised: February 19, 2009
Accepted: February 26, 2009
Published online: March 28, 2009
AIM: To investigate the effects of mutations in domain III of the hepatitis C virus (HCV) internal ribosome entry sequences (IRES) on the response of chronic HCV genotype 4a patients to interferon therapy.
METHODS: HCV RNA was extracted from 19 chronic HCV 4a patients receiving interferon/ribavirin therapy who showed dramatic differences in their response to combination therapy after initial viral clearance. IRES domain III was cloned and 15 clones for each patient were sequenced. The obtained sequences were aligned with genotype 4a prototype using the ClustalW program and mutations scored. Prediction of stem-loop secondary structure and thermodynamic stability of the major quasispecies in each patient was performed using the MFOLD 3.2 program with Turner energies and selected constraints on base pairing.
RESULTS: Analysis of RNA secondary structure revealed that insertions in domain III altered Watson-Crick base pairing of stems and reduced molecular stability of RNA, which may ultimately reduce binding affinity to ribosomal proteins. Insertion mutations in domain III were statistically more prevalent in sustained viral response patients (SVR, n = 14) as compared to breakthrough (BT, n = 5) patients.
CONCLUSION: The influence of mutations within domain III on the response of HCV patients to combination therapy depends primarily on the position, but not the frequency, of these mutations within IRES domain III.
- Citation: El Awady MK, Azzazy HM, Fahmy AM, Shawky SM, Badreldin NG, Yossef SS, Omran MH, Zekri ARN, Goueli SA. Positional effect of mutations in 5'UTR of hepatitis C virus 4a on patients' response to therapy. World J Gastroenterol 2009; 15(12): 1480-1486
- URL: https://www.wjgnet.com/1007-9327/full/v15/i12/1480.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.1480
The hepatitis C virus (HCV) genome is a 9.5 Kb single stranded RNA molecule of positive polarity. It contains a single open reading frame flanked by 5' and 3' untranslated regions (UTR) of 341 and about 230 nucleotides, respectively[1]. Both the 5' and 3' UTRs contain conserved RNA structures essential for polyprotein translation and genome replication[2]. The 5'UTR is uncapped and contains the internal ribosomal entry site (IRES) which contains highly conserved secondary and tertiary structures that are essential for proper binding and positioning of the viral RNA within the host cell's protein translation machinery[34].
The only FDA approved medication for the treatment of HCV infection is the combination therapy of either standard or pegylated interferon and ribavirin[5]. However, only 28%-60% of HCV patients respond to the treatment, depending on the viral genotype[67]. Resistance to interferon therapy is believed to be controlled by host and viral factors[8]; a significant viral factor is the generation of different HCV quasispecies[9]. The critical role of 5'UTR in initiation of polyprotein translation requires the highest degree of conservation of this region. It contains four highly structured domains numbered I to IV; all are required for recruiting, positioning, and activating/regulating the host protein synthesis machinery[10]. Translation initiation in HCV starts by binding of the HCV IRES to the 40S subunit and assembly of this binary complex with eukaryotic initiation factor 3 (eIF3). The IRES-40S-eIF3 ternary complex combines with eIF2/GTP/initiator tRNA forming the 48S complex. Progression from 48S to 80S initiation complexes is a slow step during IRES-mediated initiation which may reflect a decrease or absence of factor activity or conformational rearrangements in the IRES leading to sub-unit joining[11]. The need for flexible rather than rigid binding allows the conformational rearrangement and subsequent efficient sub-unit joining leading to initiation of translation. Therefore low affinity binding of factors due to mutations in binding sites may be associated with more flexibility and efficient initiation of translation.
The 5'UTR can be divided into the 5' and 3' parts. The 5' part is near to a fully single stranded structure and includes domain I. The 3' part is highly structured and is essential for HCV IRES function. It folds into three additional domains II, III and IV. Domain II is a stem with several internal loops. Domain IV is a small hairpin which includes the AUG start codon. The pseudo knot joins domain II with domain III and is base-paired to the sequence directly upstream of domain IV.
The HCV IRES forms an extended structure that binds the 40S subunit by several synergetic interactions, and the domains involved in this binding have been determined by chemical and enzymatic foot-printing experiments[12]. The segment starting from nucleotide 141 to 279 which compromises domain III, the focus of the present study, is highly conserved at the primary nucleotide sequence as well as at secondary and tertiary structure levels. The basal part of domain III (including principally the pseudo knot and stem-loop IIId) includes the elements of secondary structure that determine the binding of the IRES to the 40S subunit. Besides providing affinity for the 40S subunit, the basal part of domain III is thought to be required for correct positioning of the initiation codon in the decoding center of the 40S subunit, as indicated by “toe-printing”experiments[11]. The apical part of domain III binds eIF3 via stem-loop IIIb and the four-way junction and is required for 40S and eIF3 binding.
In the present study, the positional effect of mutations on the predicted secondary structure and thermodynamic stability of domain III was examined in 19 chronic HCV type 4a patients with initial virological response defined as undetectable viremia after 12 wk from start of treatment. Only 14 patients achieved sustained virological response (SVR) i.e. negative viremia 24 wk after end of treatment. In the remaining 5 patients who suffered virological breakthrough (BT), the predicted stem-loop structures and thermodynamic stability of domain III were compared in both pre- and post-treatment samples.
Male or female patients (n =19; 18-60 years) with chronic active hepatitis C virus infection were included in this study. Patients were negative for HBsAg and HBsAb but positive for anti-HCV and HCV-RNA by RT- PCR. All had elevated ALT and AST levels and received combined therapy of pegylated IFNα-2b (100 &mgr;g/wk) plus ribavirin (800-1000 mg/d). Patients had normal values for blood counts, other liver functions, auto immune markers, T3, T4 and TSH, renal functions, blood sugar and α-fetoprotein. None of the patients had other causes of liver disease (e.g. α1 antitrypsin deficiency, Wilson’s disease, alcoholic or decompensated liver disease, obesity-induced liver disease, drug-related liver disease), no CNS trauma, or active seizures, no ischemic cardiovascular disease within the last six months or hemochromatosis. None was co-infected with HBV or schistosomiasis.
RNA extraction and reverse transcription-PCR of HCV RNA, using Qiagen single step RT-PCR kit (Qiagen, Inc., Chatsworth, CA, USA), were performed as described previously[1314]. Amplification of 266 bp was performed in a single step using primer pair; forward (nt 47-68) 5'-GTGAGGAACTACTGTCTTCACG-3' and reverse (nt 292-312) 5'-ACTCGCAAGCACCCTATC AGG-3'. Cloning of amplified products was done with TA cloning kit (Invitrogen Co., Carlsbad, CA). Fifteen clones from each subject were sequenced using the TRUGENE HCV 5-NC genotyping kit, Visible Genetics, Inc. (Toronto, Ontario, Canada) in conjunction with the Open Gene DNA sequencing system. The insert DNA was sequenced by CLIP sequencing which allows both directions of the target amplicon to be sequenced simultaneously in the same tube using two different dye labeled primers (Cy5.0 and Cy5.5) for each reaction. This method provides sequence information for both positive and negative DNA strands from a single reaction. The obtained sequences were then aligned with genotype 4a prototype using the ClustalW program (http://www.ch.embnet.org/software/clustalw.html). Among the 15 domain III sequences obtained for each patient only one sequence was found in the majority of the clones, thus representing the major quasispecies of domain III in each patient. Sequence diversities in each major quasispecies were compared with genotype 4a and various mutation types were scored.
Prediction of stem-loop structure and thermodynamic stability of the major quasispecies in each patient was performed using the MFOLD 3.2 program with Turner energies and selected constraints on base pairing as indicated[15]. The program was run on EFN server: 1996-2008, Michael Zuker, Rensselaer Polytechnic Institute (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi).
The mean number of domain III quasispecies in pre-treatment samples of SVR and BT states was about 2 quasispecies per patient with no significant difference between both groups. The quasispecies complexity of domain III tripled in post-treatment samples of BT patients. All patients included in the study were genotyped as 4a except for one SVR patient infected with a hybrid 4a/1b genotype.
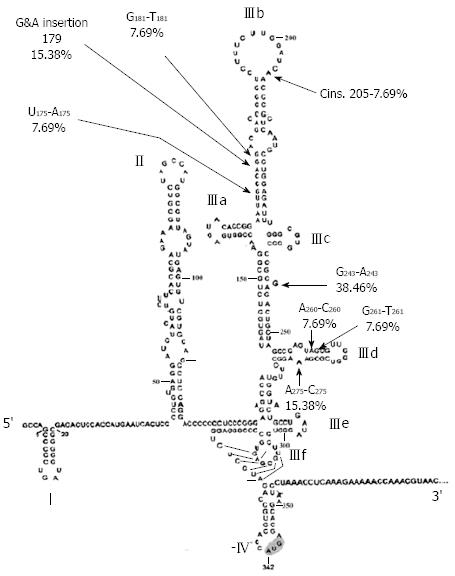
Different types of mutations were compared in both study groups with the prototype sequence of HCV 4a (Table 1). Insertion mutations in domain III were statistically more prevalent in SVR than BT patients. This is expected since insertions are known to dramatically alter the stem-loop structure by changing the Watson-Crick base pairing of stems, thus reducing molecular stability of the RNA or binding affinity to ribosomal proteins. The percentage of transition mutations was statistically higher in pre-treatment samples of BT than SVR. There were no statistical differences in percentage of transversion mutations between the two patient groups.
| Mutation | Position | Region | No. of patients | Effect on ΔG (kcal/mol) | ||
| SVR (n = 14) | BT-PreT (n = 5) | BT Post-T (n = 5) | ||||
| C-T | 148 | Junction joining stem-loop IIIa b, c & d | - | - | 1 | Increased |
| G-T | 158 | Loop IIIa | - | 1 | 4 | Increased |
| T-A | 175 | Loop IIIb | 1 | - | - | No effect |
| G & A ins | 179 | 2 | - | - | Increased | |
| T ins. | 180 | 1 | - | - | Increased | |
| T ins. | 181 | 1 | - | - | Increased | |
| C-T | 186 | - | - | 1 | No effect | |
| TTT-GGG | 194-196 | - | 1 | - | No effect | |
| C ins. | 205 | 1 | - | - | No effect | |
| T-C | 199 | - | - | 1 | No effect | |
| G-A | 243 | Junction joining stem-loop IIIa, b, c & d | 5 | 1 | 4 | No effect |
| C-T | 254 | Loop IIId | - | 1 | 1 | Decreased |
| A-C | 260 | 1 | - | 1 | Decreased | |
| A-T | 260 | 1 | - | - | Decreased | |
| G-T | 261 | 1 | - | - | Increased | |
| G-T | 268 | - | 1 | 1 | Decreased | |
| T-C | 269 | - | 1 | 1 | Decreased | |
| G ins. | 270 | - | 1 | 1 | Decreased | |
| A-C | 275 | 2 | - | - | Decreased | |
Alignment of domain III sequences derived from study patients revealed the presence of 19 point mutations scattered in stem-loops IIIa, b, c and d (Table 1). Percentage distribution of the identified mutations in different loops of domain III in SVR and BT patients is presented in Table 2. The majority of mutations, 14/19 (73.7 %), were simple substitutions while 5 (26.3%) were insertions. Substitutions included 42.8% transitions and 57.2% transversions. Most of the mutations (84.2%) were localized in 2 stem-loops; IIIb (nt 175-205) and IIId (nt 254-275), with 42.1% of the mutations in each loop. The remaining 15.8% of mutations were located in junction IIIa, b, c (10.5%) and in loop IIIa (5.3%). The SVR patients contain specific mutations (Figure 1) that were not detected in either pre-treatment BT groups or post-treatment BT patients (Table 1). These SVR specific mutations comprise 42.1% (8/19) of the total number of mutations detected in the studied patient population (Table 1). Interestingly, approximately two thirds of SVR (5/8) specific mutations were located in loop IIIb and one third (3/8) in IIId with only one exceptional mutational event (nt 243 in junction joining loops IIIa, b, c, d) that was detected outside loops IIIb and IIId. These two stem-loop structures play critical roles in directing viral protein translation via recruiting and regulating ribosomal subunit proteins and cellular initiation factors for accurate positioning of HCV RNA in the translational machinery of the host cell. On the other hand, mutational events in pre-treatment samples of the BT patients were detected in almost all loop structures of domain III, mostly located in loop IIId. Most notably loop IIId mutations were associated with decreased ΔG value (Table 1) indicating increased thermodynamic stability of the RNA structure, thus explaining resistance to therapy in the BT patient group. It is noteworthy that most of loop IIIb mutations in SVR were associated with increased ΔG value, indicating decreased thermodynamic stability of the viral RNA, therefore contributing to the multifactorial eradication of viral RNA in the SVR patient group. When comparing the mutational events in pre-treatment BT patients with those observed post-treatment, the number of mutational events known to induce significant elevation in thermodynamic stability (i.e. decreased ΔG in loop IIId) did not increase after treatment in BT patients. These results suggest that domain III-associated factors of viral breakthrough are determined by genetic events in the HCV genome before start of treatment rather than being acquired as a result of stress induced by IFNα therapy.
| Region | Responder (SVR; %) | BT pre treatment (%) | BT post treatment (%) |
| Stem-loop IIIa (156-171) | - | 14.3 | 10 |
| Stem-loop IIIb (172-227) | 50 | 14.3 | 20 |
| Stem-loop IIIc (228-238) | - | - | - |
| Stem-loop IIId (253-279) | 40 | 57.1 | 50 |
| Junction joining stem-loop IIIa, b, c, & d (141-153) & (239-252) | 10 | 14.3 | 20 |
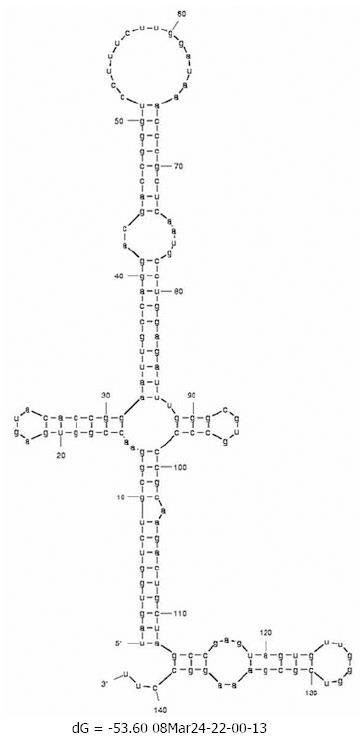
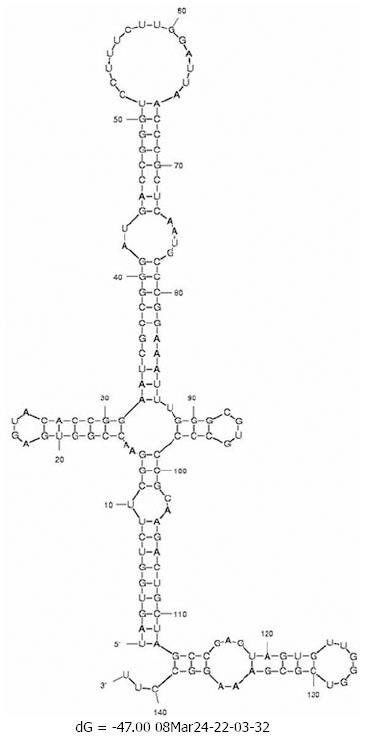
Changes in the minimum free energy (ΔG) were detected for stem-loops IIIa, b, c and d spanning nucleotides 141 to 282. Predictive stability values were compared for genotype 1b (strain H77), genotype 4a (I.D. Y11604.1) and study samples derived from SVR and BT patients. The ΔG value of genotype 1b was -53.6 kcal/mol; more stable than genotype 4a (ΔG -47.0 kcal/mol) as illustrated in the secondary structure/thermodynamic stability prediction (Figure 2 and Figure 3). As shown in Table 1, 31.5% of the mutations have no effect on the ΔG value, 31.5% of the mutations were associated with increased ΔG i.e. reduced stability of the secondary structure, while 37% of the mutations were associated with decreased ΔG value i.e. increased stability of the RNA secondary structure. An interesting observation (Table 1 and Figure 1) is that mutations with no effect on ΔG values were located in the apical part of domain III secondary structure (nt 186-243), spanning mostly the upper part of loop IIIb involved in eIF3 binding. On the other hand, those mutations affecting the stability parameters were localized in the basal part of domain III encompassing loops IIIa/b and IIId. Most notably, IIIa/b (nt 158-199) mutations were associated with reduced stability and IIId (nt 254-275) mutations were associated with increased stability of the RNA structure. Taken together, the data relating to minimum free energy in SVR and BT patients indicate that the levels of thermodynamic stability are not sufficient parameters to predict response to IFNα treatment and suggest that other parameters involving affinity of RNA binding to ribosomal subunits play important roles in determining response to treatment.
The pathway of HCV IRES-mediated initiation of translation is primarily dependent on RNA binding to several cellular proteins, of which ribosomal 40S subunit and eIF3 play pivotal roles in efficient translation of a polyprotein precursor[16]. Several foot-printing, toe-printing and UV cross linking assays have shown that domain III is the most active part in RNA-protein binding where it folds to form stem-loop structures for high affinity binding to host proteins[1117–19]. Although genomic variability in HCV IRES was shown as one mechanism for escaping IFNα effects, there has been controversy with regard to the predictive value of pre-treatment IRES genomic variations in determining later response to IFNα[20]. Although domain III harbors only 22% of the overall IRES mutations in a mixed genotype population[21], IRES activity seems to depend more on the location of mutant nucleotides which play the most important roles in IRES activity.
We performed a focused study on domain III derived from patients infected exclusively with genotype 4a and presenting a dramatic difference in sustained response after initial viral disappearance. This approach allowed us to minimize quasispecies complexity (mean of 2 variants/patient) compared with 7 or more variants/patient in other studies[21] and to pinpoint a number of important genomic determinants of response to IFNα. Cloning of domain III and sequencing of 15 clones in each patient allowed us to identify the major variant (identical sequences in 12 clones or more) in each patient so that noise was reduced during extrapolation of the relationship between genomic variation and treatment outcome.
Disruption of base pairing in stem structure significantly inhibits IRES-dependent translation[22]. In our patient population insertion mutations in domain III were statistically higher in SVR than in BT patients. These results are expected since insertions are known to dramatically alter the stem-loop structure by changing the Watson-Crick base pairing of stems, thus reducing molecular stability of the RNA or binding affinity to ribosomal proteins. The distributions of domain III mutations in SVR patients were distinct from those of the BT group in the present study. Of the total number of mutations, approximately 42% were exclusive to SVR patients; almost all were located in loops IIIb and IIId. These two stem-loop structures were implicated in initiation complex recruitment, positioning and regulation, where IIId forms the core of high affinity binding with the 40S subunit[22] and recruits ribosomal elements involved in positioning of mRNA and tRNA[10]. Besides binding to eIF3, stem-loop IIIb interacts with a multisubunit initiation factor involved in subunit assembly and stability of ternary complex[22].
This explanation cannot simply be taken to resolve mysteries of HCV IRES in light of the apparent controversy in loop IIId genomic diversity. All of IIId mutations detected exclusively in BT patients (pre- and post-treatment) were associated with increased thermodynamic stability, thus leading to viral persistence; also most of IIId mutations detected in SVR patients were again associated with increased stability of this RNA. The slow rate of ribosomal subunit transition compared to canonical translation[11] directs the attention towards the need for flexible rather than rigid binding which allows the conformational rearrangement and subsequent efficient subunit joining, leading to initiation of translation. Therefore mutations affecting binding affinity of factors regardless of thermodynamic stability of RNA structure may be associated with either more flexibility or rigidity which in turn regulates efficiency of translational initiation[11]. Taken together, the data regarding minimum free energy in SVR and BT patients indicate that the levels of thermodynamic stability are not sufficient parameters to predict response to IFNα treatment and suggest that other parameters involving affinity of RNA binding to ribosomal subunits play significant roles in determining response to treatment. The concept that mutations in BT patients appear only in post-treatment samples and are associated with no effect on RNA stability suggests that viral breakthrough is determined by mutations in domain III before start of treatment rather than being acquired during treatment. Alternatively, the roles of these mutations in viral persistence could be related to fine tuning of the flexibility of RNA structure for binding to cellular factors regardless of its stability. The former view is more plausible since the majority of IIId mutations in pre-treatment were associated with increased RNA stability without change in the frequency of mutations post-treatment. An interesting observation in this study is that the A243G mutation in the IIIc/IIId junction was detected both in SVR (5 times) and in pre-treatment BT (2 times) and was more detectable in post-treatment BT patients (4 times). Predictive folding however, revealed no effect of this mutation on the calculated thermodynamic stability. The role of nucleotide 243 in maintaining IRES structure was reported in HCV genotype 1b[21] and changes at this position were encountered in patients with viral stabilization. In genotype 1b, A243 pairs with U149, which is lacking in 4a, leading to altered pairing and explains the high rate of mutations at this position in our study population regardless of response to IFNα, thus making it more vulnerable to mutational event in genotype 4a.
In conclusion, the RNA structure of domain III in HCV IRES contains several important elements implicated in determining the response to IFNα treatment. The results presented herein demonstrate that domain III structure in SVR patients is different from BT patients. Thermodynamic stability of RNA secondary structure is a significant but not sufficient parameter for prediction of viral stabilization, or response to IFNα. Elements of binding to ribosomal subunit complexes require further studies to unravel the exact role of IRES in HCV stabilization and persistence.
The hepatitis C virus (HCV) is a major public health problem with about 200 million individuals currently infected with the virus (about 3% of the world’s population). So far, 11 genotypes and more than 70 subtypes have been identified. The only approved FDA treatment for chronic HCV infection is the combination therapy of pegylated interferon and ribavarin. The variable response of patients to therapy ranges between 28% and 60% and has been proposed to be affected by various host and viral factors. Investigating the effect of mutations within the HCV 5’UTR, the most conserved region in the viral genome, on response to therapy is important because this region is vital for initiation of viral polyprotein translation and the ability of HCV to replicate.
Although interferon and ribavirin are the only FDA approved drugs to treat HCV, they suffer from several drawbacks including severe side effects (including hematological abnormalities and neuropsychiatric symptoms), very high cost, and most importantly low therapeutic response. Consequently, factors that affect the response of HCV patients to therapy have to be addressed and results could be used for predicting response to therapy before initiation of treatment. Moreover, identification of viral factors that correlate with therapeutic response would contribute to other studies on viral and host factors. This could result in a global view and comprehensive understanding of how host and viral factors affect a patient’s response to therapy.
Recent reports, using clinical specimens or HCV replicon systems from different genotypes, have highlighted the effect of mutations in different domains of the viral genome (in particular the HCV 5’UTR) on patients’ response to therapy. However, limited studies have been done on genotype 4a. In the present article, The authors focused mainly on genotype 4a which is the predominant genotype in Egypt; found in over 90% of all HCV-infected patients. They showed that the thermodynamic stability of the HCV 5’UTR region is different among responders (sustained viral clearance) and breakthrough patients (who suffer relapse at the end of treatment). Additionally, their results indicate that response to therapy is related mainly to the position of mutations but not their frequency. Finally, thermodynamic stability of IRES was shown to have a direct influence on the binding of the viral genome to the host proteins, which results in initiation of the translation of the viral polyprotein.
The results of this study suggest that the presence of single nucleotide polymorphisms (SNPs) in certain positions had direct effect on the response of HCV patients to interferon therapy. Taking into consideration the positions of these mutations, different real-time PCR or other assays can be developed for detection of the SNPs to allow the prediction of the response to interferon therapy as a step for identification of patients who are more likely to respond to therapy.
5’UTR: non-coding region of HCV RNA; contains the internal ribosomal entry site (IRES); site of initiation of translation. IRES: (internal ribosomal entry site): a structure within the HCV RNA 5’UTR that binds directly to the ribosome to initiate translation. Cap-dependent translation: the mechanism of translation (protein synthesis) predominantly used for cellular proteins. Cap-independent translation: translation via an internal ribosomal entry site (IRES); the mechanism utilized by HCV. Eukaryotic initiation factors: cellular proteins involved in translation. SVR: sustained viral response; patients who show negative HCV PCR results after termination of therapy.
The authors present a sequence analysis study of HCV genotype 4a patients undergoing combination therapy. This study is of a substantial potential interest. In this work the authors propose that insertion mutations in domain III of the IRES region are more prevalent in sustained viral response patients compared with breakthrough patients and that such mutations may affect the ability of HCV virus to replicate by decreasing the thermodynamic stability of its RNA.
| 1. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [Cited in This Article: ] |
| 2. | Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5-19. [Cited in This Article: ] |
| 3. | Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5' border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165-1174. [Cited in This Article: ] |
| 4. | Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol. 2002;9:370-374. [Cited in This Article: ] |
| 5. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [Cited in This Article: ] |
| 6. | Booth JC, O'Grady J, Neuberger J. Clinical guidelines on the management of hepatitis C. Gut. 2001;49 Suppl 1:I1-21. [Cited in This Article: ] |
| 7. | Hofmann WP, Zeuzem S, Sarrazin C. Hepatitis C virus-related resistance mechanisms to interferon alpha-based antiviral therapy. J Clin Virol. 2005;32:86-91. [Cited in This Article: ] |
| 8. | Yeh SH, Chen DS, Chen PJ. A prospect for pharmaco-genomics in the interferon therapy of chronic viral hepatitis. J Antimicrob Chemother. 2003;52:149-151. [Cited in This Article: ] |
| 9. | Salmeron J, Casado J, Rueda PM, Lafuente V, Diago M, Romero-Gomez M, Palacios A, Leon J, Gila A, Quiles R. Quasispecies as predictive factor of rapid, early and sustained virological responses in chronic hepatitis C, genotype 1, treated with peginterferon-ribavirin. J Clin Virol. 2008;41:264-269. [Cited in This Article: ] |
| 10. | Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101:16990-16995. [Cited in This Article: ] |
| 11. | Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369-380. [Cited in This Article: ] |
| 12. | Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194-206. [Cited in This Article: ] |
| 13. | Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495-503. [Cited in This Article: ] |
| 14. | Zekri AR, El-Din HM, Bahnassy AA, Khaled MM, Omar A, Fouad I, El-Hefnewi M, Thakeb F, El-Awady M. Genetic distance and heterogenecity between quasispecies is a critical predictor to IFN response in Egyptian patients with HCV genotype-4. Virol J. 2007;4:16. [Cited in This Article: ] |
| 15. | Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406-3415. [Cited in This Article: ] |
| 16. | Dasgupta A, Das S, Izumi R, Venkatesan A, Barat B. Targeting internal ribosome entry site (IRES)-mediated translation to block hepatitis C and other RNA viruses. FEMS Microbiol Lett. 2004;234:189-199. [Cited in This Article: ] |
| 17. | Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5' nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775-4782. [Cited in This Article: ] |
| 18. | Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67-83. [Cited in This Article: ] |
| 19. | Buratti E, Tisminetzky S, Zotti M, Baralle FE. Functional analysis of the interaction between HCV 5'UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 1998;26:3179-3187. [Cited in This Article: ] |
| 20. | Yamamoto C, Enomoto N, Kurosaki M, Yu SH, Tazawa J, Izumi N, Marumo F, Sato C. Nucleotide sequence variations in the internal ribosome entry site of hepatitis C virus-1b: no association with efficacy of interferon therapy or serum HCV-RNA levels. Hepatology. 1997;26:1616-1620. [Cited in This Article: ] |
| 21. | Thelu MA, Leroy V, Ramzan M, Dufeu-Duchesne T, Marche P, Zarski JP. IRES complexity before IFN-alpha treatment and evolution of the viral load at the early stage of treatment in peripheral blood mononuclear cells from chronic hepatitis C patients. J Med Virol. 2007;79:242-253. [Cited in This Article: ] |