INTRODUCTION
Probiotics are defined as living microorganisms that have beneficial effects on human health[1]. Documented health effects in human intervention trials include amelioration of acute diarrhoea in children, reduction of the risk of antibiotic-associated gastrointestinal symptoms, relief of milk allergy/atopic dermatitis in infants, reduction in the risk of atopic diseases and respiratory infections, relief of irritable bowel syndrome and rheumatoid arthritis symptoms, suppression of H pylori and modulation of the immune response[2–4]. The ways in which probiotic bacteria affect the human immune system in vivo and in vitro are not fully understood. The immunomodulatory effects of probiotic bacteria could be due to the produced cytokines that further regulate innate and adaptive immune responses.
Different leukocyte types cooperate during the activation of innate and adaptive immune responses. In addition to direct cellular contacts the communication between immune cells depends on secreted mediators including cytokines. Bacteria-induced stress produces inflammatory cytokines promoting the activation of antimicrobial immune responses. TNF-α is a potent inducer of many inflammatory molecules including other cytokines[5]. IL-12 produced by activated antigen-presenting cells (APC) enhances the development of Th1 type immune responses and stimulates NK and T cell IFN-γ production. All this further enhances cell-mediated Th1 type responses[6]. IL-10 downregulates the inflammatory response and induces an antibody-mediated immune response[7]. In previous studies it has been shown that individual probiotic bacteria can induce the production of TNF-α[8–10], IL-12[11–15], IFN-γ[10111315] and IL-10[915–18] in human peripheral blood mononuclear cells (PBMC) in vitro. At present only a limited amount of comparative data is available on the ability of different probiotic strains to induce cytokine production within the same experimental system[914–17]. Also, the effect of probiotic bacterial combinations on cytokine production in vitro is not well documented.
Understanding of the cytokine patterns that probiotics elicit may help in designing probiotics for specific preventative or therapeutic purposes. Information on the cytokine production induced by different probiotic bacteria and their combinations would enable development and optimal clinical use of these microbes as health promoting substances. In the present study human PBMCs consisting of monocytes, T and B lymphocytes and NK cells, are used for in vitro screening for cytokine production. Cytokine production in response to stimulation with different potentially probiotic strains from six bacterial genera Streptococcus, Lactobacillus, Bifidobacterium, Lactococcus, Leuconostoc and Propionibacterium alone or in combinations is analysed in order to identify potential enhancing or synergistic effects.
MATERIALS AND METHODS
Bacterial strains
Streptococcus pyogenes serotype T1M1 (IH32030) and Escherichia coli (DH5α) were obtained from the collection of National Public Health Institute (Helsinki, Finland). Eleven potentially probiotic strains; Streptococcus thermophilus THS, Lactobacillus rhamnosus GG (ATCC 53103), Lactobacillus rhamnosus Lc705 (DSM 7061), Lactobacillus helveticus 1129, Lactobacillus helveticus Lb 161, Bifidobacterium longum 1/10, Bifidobacterium animalis ssp. lactis Bb12, Bifidobacterium breve Bb99 (DSM 13692), Lactococcus lactis ssp. cremoris ARH74 (DSM 18891), Leuconostoc mesenteroides ssp. cremoris PIA2 (DSM 18892) and Propionibacterium freudenreichii ssp. shermanii JS (DSM 7067), were obtained from Valio Research Centre (Helsinki, Finland). Bacteria were stored in skimmed milk at -70°C and passaged three times (except Bifibobacterium strains which were passaged four times) before they were used in stimulation experiments. S. pyogenes was grown at 37°C under aerobic conditions in sheep blood agar (Oxoid, Ogdensburg, NY, USA) and tryptone-yeast broth supplemented with 2 g/L glucose[19], E. coli at 37°C under aerobic conditions in Luria-medium (National Public Health Institute, Helsinki, Finland), Lactobacillus rhamnosus strains were grown at 37°C under aerobic conditions in de Man, Rogosa and Sharpe (MRS) medium (Lab M, Topley House, Lancashire, UK), Lactobacillus helveticus strains were grown at 42°C under aerobic conditions in MRS medium (Lab M, Topley House), L. mesenteroides was grown at 22°C under aerobic conditions in MRS medium (Lab M, Topley House), Bifidobacterium strains at 37°C under anaerobic conditions in MRS medium (Lab M, Topley House) with 5 g/L cysteine (Merck, Darmstadt, Germany), S. thermophilus at 37°C under aerobic conditions in M17-agar (Lab M, Topley House) with 20 g/L lactose (J.T. Baker B.V., Deventer, Holland) and M17-broth (Difco, Beckton Dickinson, MD, USA) with 20 g/L lactose (J.T. Baker B.V.), P. freudenreichii at 30°C under aerobic conditions in propioni-medium (Valio Ltd, Helsinki, Finland), L. lactis at 22°C under aerobic conditions in calcium citrate agar (Valio Ltd) and M17-broth (Difco) with 20 g/L lactose (J.T. Baker B.V.). For stimulation experiments bacteria were grown to logarithmic growth phase, and the number of bacteria was determined by counting in a Petroff-Hauser counting chamber.
Cell culture
Human PBMC were purified by density gradient centrifugation over a Ficoll-Paque gradient (Amersham-Pharmacia Biotech, Uppsala, Sweden) from freshly collected, leukocyte-rich buffy coats obtained from healthy blood donors (Finnish Red Cross Blood Transfusion Service, Helsinki, Finland)[20]. After washing, the cells were resuspended in RPMI 1640 medium (Sigma, St. Louis, Mo., USA) containing 10% heat-inactivated fetal calf serum (Integro, Zaandam, Holland) and supplemented with 2 mmol/L L-glutamine (Sigma), 100 U/mL penicillin and 100 mg/mL streptomycin (Gibco BRL, Paisley, Scotland). In stimulation experiments purified leukocytes (2 × 109 cells/mL) were incubated with bacteria in a final volume of 1 mL in 24-well plates (Nunc, Roskilde, Denmark) in 5% CO2 at 37°C.
Stimulation experiments
All experiments were performed with cells obtained from four different blood donors. During bacterial stimulations PBMCs were maintained in RPMI-1640 medium containing 100 mL/L FCS. Bacteria were added into the cell culture to obtain the required bacteria: host cell ratio. S. pyogenes was used as a positive control and RPMI 1640 containing 100 mL/L FCS as a negative control. Bacterial doses and incubation times are as indicated for each experiment. When PBMC were stimulated with a combination of two bacteria or more, equal numbers of different bacteria were used, and the sum bacterial dose of the combinations was 10:1 of a bacteria: host cell ratio. Cell culture supernatants were collected from individual donor cell cultures and stored at -20°C before analysis. For RNA analysis cells from different donors were pooled.
Cytokine specific ELISA
Cell culture supernatants obtained from individual donors were analyzed for cytokine levels by using enzyme-linked immunosorbent assay (ELISA) essentially as previously described[8]. TNF-α and IL-10 were determined with antibody pairs and standards obtained from BD Pharmingen (San Diego, CA, USA). IFN-γ and IL-12p70 were determined with Eli-pair kits (BioSite, Täby, Sweden).
RNA isolation and Northern blotting
For isolation of total cellular RNA, stimulated cells from different donors were pooled, collected, washed with PBS, and lysed in guanidinium isothiocyanate[21], followed by a centrifugation through a CsCl cushion as previously described[22]. RNA was quantified photometrically and samples containing equal amounts (10 &mgr;g) of total cellular RNA were size-fractioned on 1% formaldehyde-agarose gels, transferred to Hybond-N nylon membranes (Amersham-Pharmacia-Biotech) and hybridized. To control equal loading, ethidium bromide staining was used. The cDNA probes were human TNF-α (ATCC), IL-12 p40 and p35[23], IFN-γ[24] and IL-10 (DNAX, Palo Alto, USA). Hybridizations were performed in a solution containing 500 g/L formamide, 5 × Denhardt’s solution, 5 × SSPE and 5 g/L SDS at 42°C. After hybridization membranes were washed three times with 1 × saline sodium citrate/g per L SDS at 42°C for 30 min and once at 65°C for 30 min. Membranes were exposed to Kodak X-Omat AR films (Eastman Kodak, Rochester, NY, USA) at -70°C with intensifying screens.
RESULTS
Bacterial dose-dependent induction of cytokine production in human PBMC
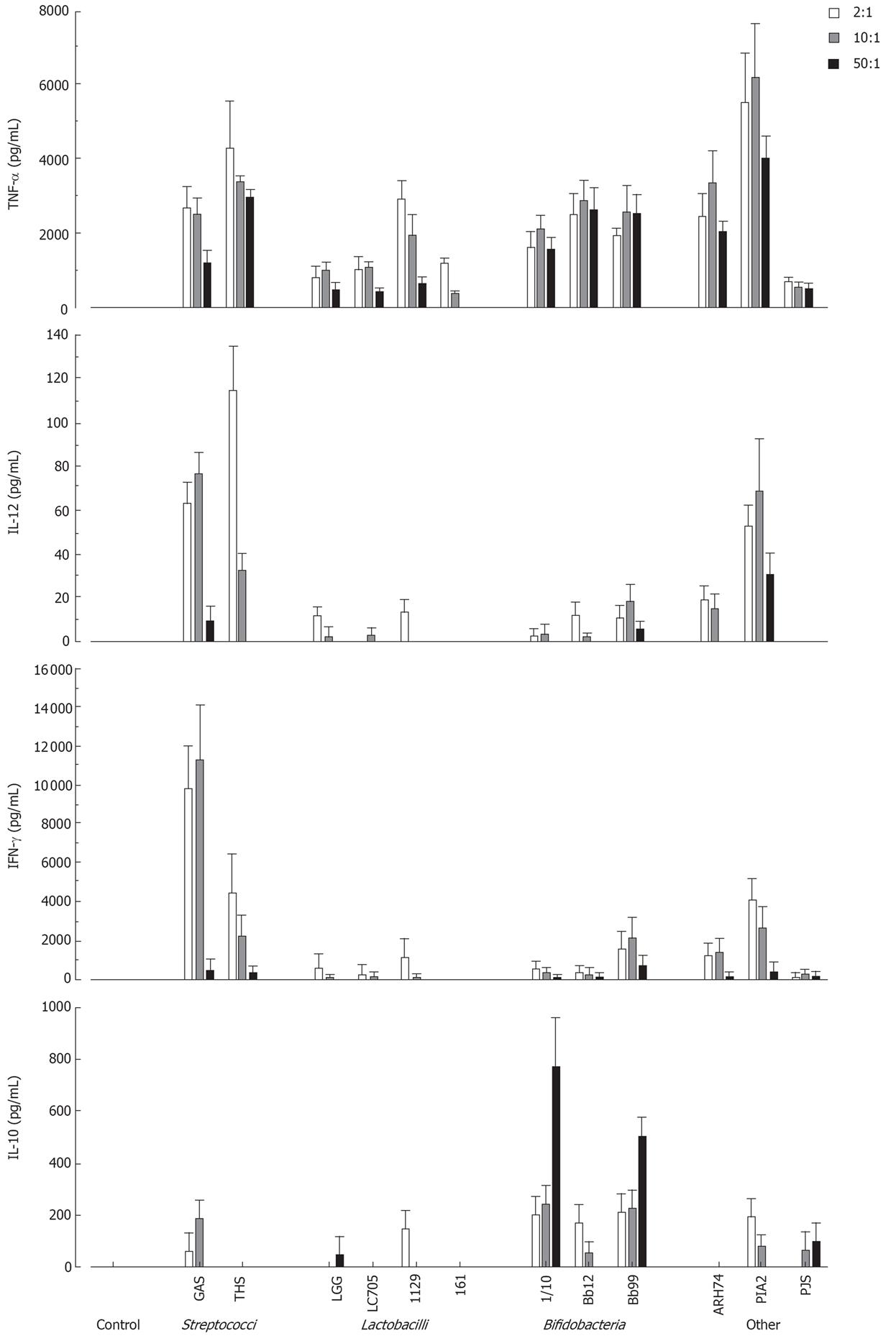
To determine the optimal bacterial dose that can induce cytokine production in human PBMC, cells were stimulated with different doses of live probiotic bacteria (2:1, 10:1 and 50:1 bacteria: cell ratio). Cell culture supernatants were collected at 24 h after stimulation and cytokine levels were determined by ELISA. As shown in Figure 1 all bacteria induced TNF-α production and L. mesenteroides was the most potent inducer. IL-12 and IFN-γ were best induced by streptococci and L. mesenteroides. Bifidobacteria induced IL-10 production in a dose-dependent manner, while other probiotic species were weak inducers of IL-10 production (Figure 1). Bacteria: host cell ratio of 10:1 was chosen for more detailed analyses.
Figure 1 Dose-dependent cytokine production in human PBMC (2 × 106/mL) stimulated with Streptococcus pyogenes and 11 probiotic bacteria (2, 10 and 50 bacteria/cell) for 24 h, mean ± SEM, n = 4.
A representative experiment out of two is shown. GAS: Streptococcus pyogenes; THS: Streptococcus thermophilus THS; LGG: Lactobacillus rhamnosus GG; LC705, Lactobacillus rhamnosus LC705; 1129: Lactobacillus helveticus 1129; 161: Lactobacillus helveticus Lb 161; 1/10: Bifidobacterium longum 1/10; Bb12: Bifidobacterium animalis ssp. lactis Bb12; Bb99, Bifidobacterium breve Bb99; ARH74: Lactococcus lactis ssp. cremoris ARH74; PIA2, Leuconostoc mesenteroides ssp. cremoris PIA2; PJS: Propionibacterium freudenreichii ssp. shermanii JS.
Kinetics of cytokine production in probiotic bacteria-stimulated PBMC
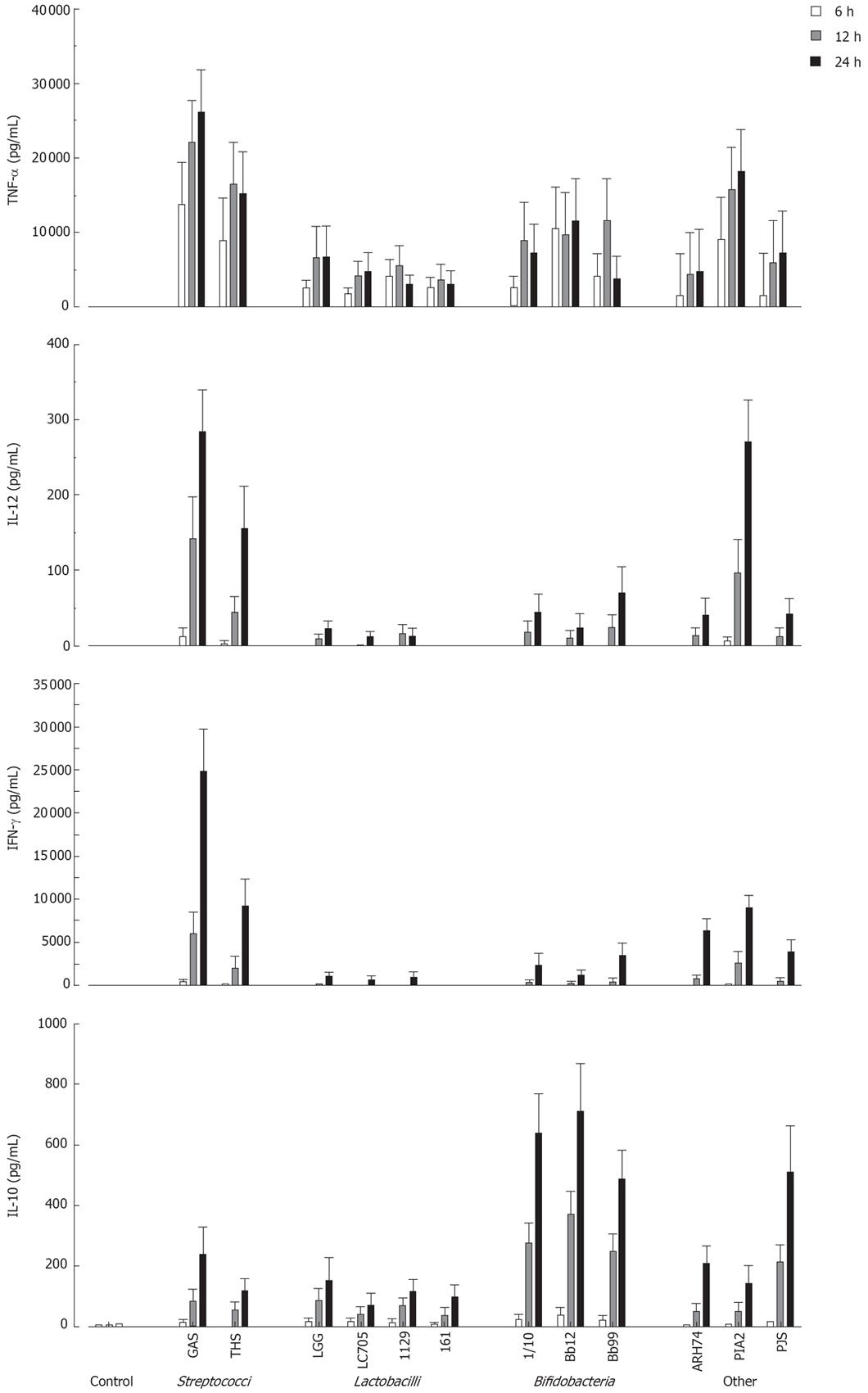
Next the kinetics of cytokine production in PBMC using a 10:1 bacterium: host cell ratio was analysed by ELISA. All probiotic bacteria induced TNF-α production by 6 h after stimulation (Figure 2). With S. thermophilus, Lactobacillus strains, B. longum and B. breve maximal TNF-α production levels were reached at 12 h after stimulation (Figure 2). Other measured cytokines were secreted later, and all tested probiotic bacteria induced maximal IL-12, IFN-γ and IL-10 production at 24 h after stimulation (Figure 2). There were considerable differences in the ability of studied bacteria to induce cytokine production in PBMC. S. thermophilus and L. mesenteroides were the best inducers of proinflammatory cytokines TNF-α, IL-12 and IFN-γ. Anti-inflammatory IL-10 production was induced by all Bifidobacterium strains and P. freudenreichii. All strains of Lactobacillus were weak inducers of IL-10 and Th1 type cytokines.
Figure 2 Kinetics of cytokine production in probiotic bacteria-induced human PBMC.
(10 bacteria/cell, mean ± SEM, n = 4). See Figure 1 for abbreviations of bacteria.
Probiotic bacteria-induced cytokine mRNA expression
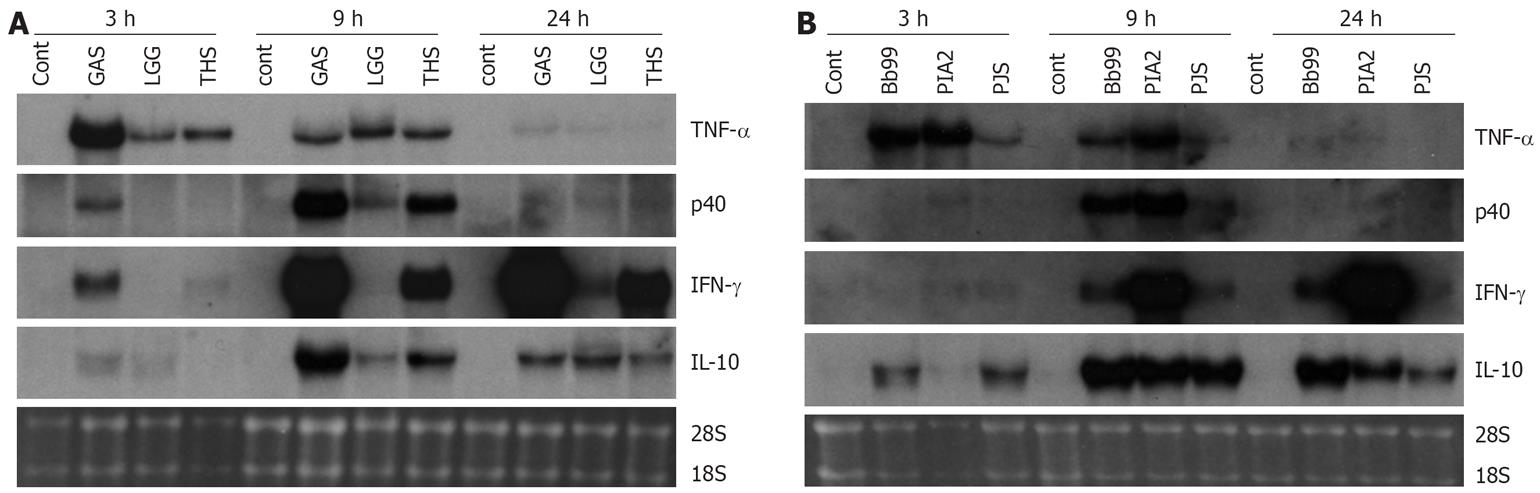
Those bacteria that were the best inducers of cytokines at the protein level (namely S. thermophilus, B. breve, L. mesenteroides and P. freudenreichii) were selected for more detailed analysis of cytokine mRNA expression as analyzed by Northern blotting. In addition to our positive control S. pyogenes, also L. rhamnosus GG that has been extensively studied in vitro by our group and in many clinical trials[23] was added for the analysis. Consistent with the kinetics of TNF-α protein production, mRNA expression was fast and detectable at 3 h after bacterial stimulation (Figure 3). With all studied bacteria, the kinetics of TNF-α mRNA expression was similar. IL-12 p40, IFN-γ and IL-10 genes were induced later than TNF-α and their expression was detectable starting at 9 h after stimulation (Figure 3). S. thermophilus, B. breve and L. mesenteroides induced IL-12 p40 mRNA expression as well as S. pyogenes, a well-known inducer of IL-12 p40 in PBMCs (Figure 3)[11]. S. thermophilus and L. mesenteroides induced IL-12 p40 mRNA expression at 9 h while P. freudenreichii induced the expression only weakly. IL-12 p35 mRNA expression remained under the detection limit (data not shown). The kinetics of IFN-γ mRNA expression differed from that of IL-12 p40. In probiotic bacteria-stimulated PBMCs IFN-γ mRNA expression was observed at 9 h after stimulation, and its expression increased until the 24 h time point. L. mesenteroides, S. thermophilus and B. breve, in this rank order were able to induce IFN-γ mRNA expression (Figure 3). IL-10 mRNA expression was highest at 9 h after bacterial stimulation persisting until the 24 h time point. Interestingly, B. breve and P. freudenreichii were able to induce IL-10 mRNA expression already at 3 h after stimulation (Figure 3). The kinetics of probiotic bacteria-induced cytokine mRNA expression was in line with the cytokine secretion (Figures 2 and 3).
Figure 3 Kinetics of cytokine mRNA expression in probiotic bacteria-induced human PBMC (pooled samples from four donors), 10 bacteria/cell, 10 &mgr;g RNA/lane on 10 g/L agarose gel and Northern blot analysis with indicated cDNA probes was carried out.
Ethidium bromide staining of gels was used to verify equal loading of RNA. See Figure 1 for abbreviations of bacteria. A: S. pyogenes, L. rhamnosus GG or S. thermophilus and B: B. breve, L. mesenteroides, P. freudenreichii.
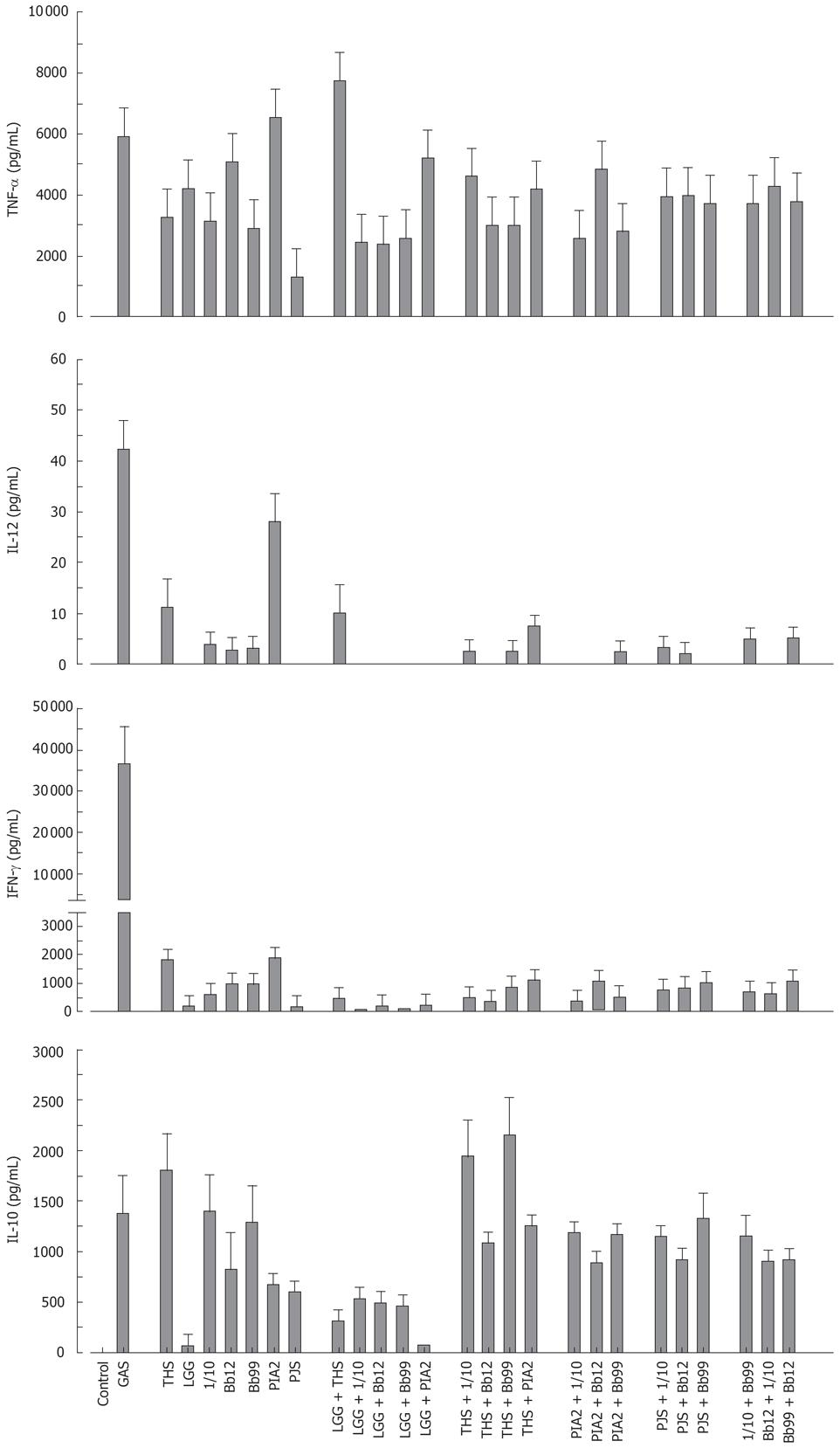
Combination effect of probiotic bacteria on cytokine production in human PBMC
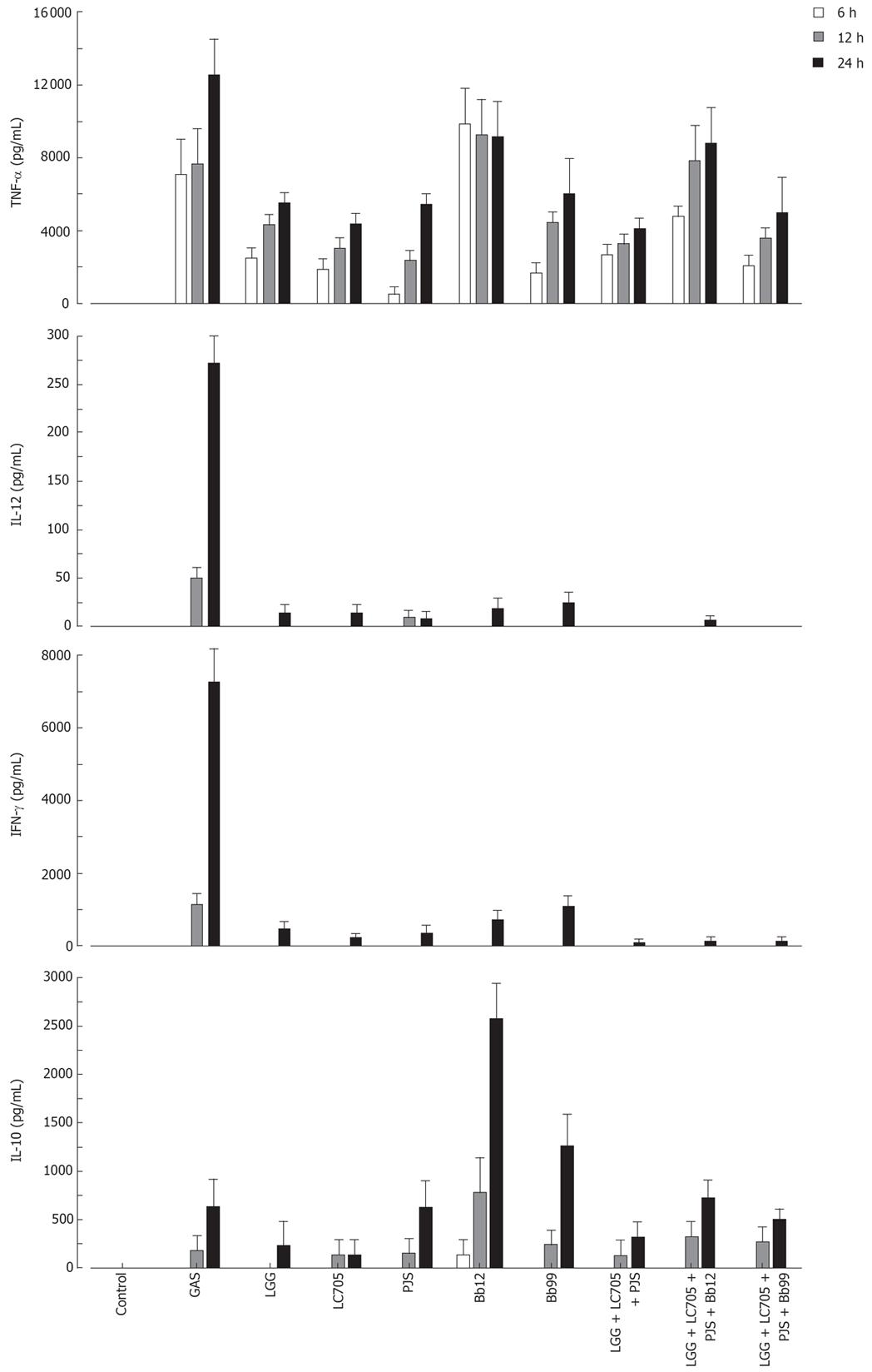
The probiotic bacteria and their combinations which have been used in clinical trials, namely L. rhamnosus strains, P. freudenreichii and B. breve[25–28], L. rhamnosus strains, P. freudenreichii, and B. animalis[2930] and L. rhamnosus strains and P. freudenreichii[31], were used to study whether the effect of probiotic combinations differs from an individual strain. All probiotic combinations induced TNF-α production at the same or lower level as compared to those responses induced by probiotic bacteria alone (Figure 4). IL-12 and IFN-γ production induced by all probiotic bacterial combinations was weak (Figure 4). Also the IL-10 production by the probiotic combinations was lower than by individual Bifidobacterium strains (Figure 4) In general; the responses induced by the combinations of probiotic bacteria were an average of responses induced by each bacterium alone (Figure 4). In addition the representatives of the best cytokine inducers in this study, namely S. thermophilus, Bifidobacterium strains, L. mesenteroides and P. freudenreichii and our controls S. pyogenes and L. rhamnosus GG were analysed for their possible additive and synergistic effects on cytokine production in PBMCs. Two different probiotic bacteria were used simultaneously to stimulate PBMCs for 24 h with a bacteria: host cell ratio of 10:1 and cell culture supernatants were collected for ELISA analysis. None of these probiotic bacterial combinations induced additive or synergistic cytokine production in PBMCs. On the contrary, the responses induced by the combinations were closer to an average of responses induced by the individual bacteria (Figure 5).
Figure 4 Kinetics of cytokine production in human PBMC stimulated with bacterial combinations.
(10 single bacteria/cell, or three bacteria/cell in the combinations, n = 4). See Figure 1 for abbreviations of bacteria.
Figure 5 Cytokine production in human PBMC stimulated with probiotic bacterial combinations.
(10 single bacteria/cell or their combinations at 5/cell for 24 h, mean ± SEM, n = 4). See Figure 1 for abbreviations of bacteria.
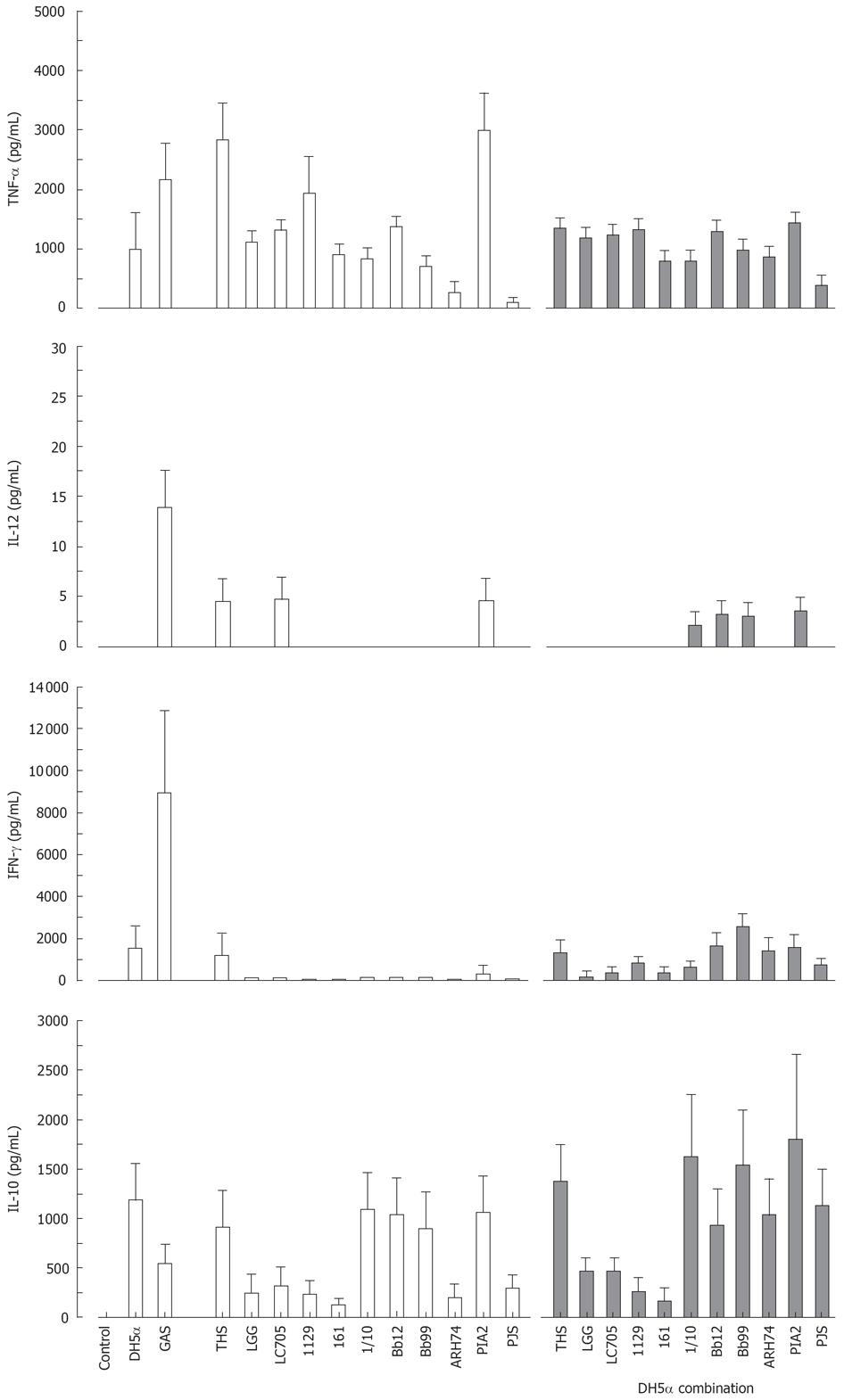
Effect of probiotic bacteria together with E. coli on cytokine production in human PBMC
All studied probiotic bacteria are gram-positive and thus likely to use Toll-like receptor (TLR)2-mediated signal transduction pathways in host cells while a gram-negative bacterium E. coli uses TLR4[32]. We did not observe any additive effects on cytokine production induced by the combinations of our gram-positive probiotic bacteria (Figure 5). Since this could be due to our probiotic bacteria engaging the same receptor, we wanted to analyse whether individual probiotic bacteria combined with E. coli would have an effect on cytokine production. Probiotic S. thermophilus, Lactobacillus, Bifidobacterium, Lactococcus, Leuconostoc or Propionibacterium strain and E. coli were combined and cell culture supernatants were collected at 24 h after bacterial stimulation followed by determination of cytokine production by ELISA. E. coli induced TNF-α, IFN-γ and IL-10 production, but not that of IL-12 (Figure 6). Stimulation of PBMCs with different combinations of probiotic bacteria together with E. coli lead to the induction of TNF-α production, but the response was an average of responses with individual bacteria (Figure 6). E. coli-induced IFN-γ production was reduced when it was combined with Lactobacillus strains, P. freudenreichii and B. longum (Figure 6). No enhancement of IL-10 production was seen in cells stimulated with the combinations of E. coli and probiotic bacteria.
Figure 6 Stimulation of PBMC with a combination of gram-positive probiotic bacteria and a gram-negative bacteria for 24 h.
(E. coli DH5α or different probiotic bacteria alone at 10 bacteria/cell, or E. coli combined with each probiotic bacterium at 5 bacteria/cells means ± SEM, n = 4). Abbreviations of bacteria see Figure 1.
DISCUSSION
In this study we have systematically analysed the ability of potentially probiotic bacterial strains from six different genera; Streptococcus, Lactobacillus, Bifidobacterium, Lactococcus, Leuconostoc and Propionibacterium, to induce cytokine expression in vitro in human PBMC. We show that eleven probiotic strains induce cytokine expression differently and the expression pattern seems to be dependent on the bacterial genera. In addition, stimulation of PBMC with any bacterial combinations, whether gram-positive or gram-negative, does not lead to enhanced cytokine production. Also we report for the first time that novel probiotic S. thermophilus and Leuconostoc strains are more potent inducers of Th1 type cytokines IL-12 and IFN-γ than the probiotic Lactobacillus strains presently in clinical use.
Human PBMC offer a model for studying the potential of different probiotic strains to induce cytokine production. In vivo, probiotics are not in direct contact with PBMC. Instead, probiotics interact with the epithelial cells of the gut, and probiotics may be taken up by macrophages, dendritic cells or M-cells at Peyer’s patches and this can lead to the activation of lymphocytes such as monocytes residing in Peyer’s patches[33]. Since PBMC are a source for monocytes among other immune cells, they provide an adequate model to study the immunological properties of probiotic bacteria. Therefore, in vitro bacteria-host cell studies aid in selecting novel probiotic strains for clinical trials.
Our data is consistent with previous studies showing that probiotic bacteria are able to induce TNF-α secretion in human PBMC[8101117]. However, there were considerable differences in the ability of different probiotic bacteria to induce IL-12 and IFN-γ. Interestingly, Lactobacillus and Bifidobacterium strains which have previously been shown to stimulate IL-12 and IFN-γ production in human PBMC[11–1417], were in the present study found to be relatively poor inducers of these cytokines. Instead, we found that novel probiotic S. thermophilus and Leuconostoc strains were extremely good inducers of these Th1 type cytokines. One of the approaches in treating allergy could be tipping the Th1/Th2 balance from Th2 predominance to Th1 type response. In clinical trials L. rhamnosus GG[273435] or a combination of two different L. rhamnosus strains, Bifidobacterium and Propionibacterium[28] have been used successfully to prevent atopic diseases. A strong Th1 type cytokine response is also an important factor in the fight against viral infections such as that caused by influenza A virus. Indeed, Lactobacillus strains have been shown to prevent and ameliorate the symptoms of respiratory infections[36–40]. It is of interest that for example L. rhamnosus GG has been effective in immune mediated diseases even though in the present study we observed that LGG induced very low cytokine production. This may be due to the fact that cytokine induction is only one of the proposed mechanisms of action for probiotics. Other as important factors may for example be the ability to adhere to the gut epithelium and the promotion of non-immunological gut defence barrier by normalizing permeability and disturbed gut microecology. Since S. thermophilus and Leuconostoc strains used in the present study were extremely potent inducers of IL-12 and IFN-γ, these strains may show better clinical efficiency in enhancing Th1 response in allergy and in the prevention of respiratory infections than the presently used probiotic Lactobacillus strains.
IL-10 was induced by Bifidobacterium and Propionibacterium strains, whereas IL-10 production induced by Streptococcus, Lactobacillus, Lactococcus and Leuconostoc strains remained at a low level. Our data is consistent with previous studies in which bifidobacteria were shown to induce higher IL-10 production as compared to lactobacilli[916]. The anti-inflammatory actions of IL-10 could be helpful in the treatment of inflammatory conditions or diseases. There is preliminary evidence that probiotics could be used in the treatment of inflammatory diseases like ulcerative colitis, pouchitis and rheumatoid arthritis[241]. The amelioration of these inflammatory diseases could be due to the induction of IL-10. Recent studies indicate a possible role of low-grade mucosal inflammation also in the pathogenesis of irritable bowel syndrome[4243]. Preliminary evidence exists that a combination of probiotics, which included anti-inflammatory Bifidobacterium and Propionibacterium strains in addition to two different L. rhamnosus strains, relieves the symptoms of irritable bowel syndrome[25]. Bifidobacterium and Propionibacterium which in this study were able to induce anti-inflammatory IL-10 production could thus be used to treat different types of inflammatory diseases.
The use of probiotic bacterial combinations in clinical trials has shown great promise making it important to understand the immunological properties of a single strain versus different bacterial combinations. However, there is only limited amount of comparative data on the immunomodulatory properties of several different probiotic bacterial strains within the same experimental system. Published data is mainly limited to the analysis of the effects of Lactobacillus and Bifidobacterium genera[911–1344]. In the present study combinations of different gram-positive probiotic bacteria did not induce any additive or synergistic cytokine production in PBMCs. This could be due to the fact that all gram-positive bacteria are likely to use the same or similar intracellular signal transduction mechanisms to induce cytokine gene expression. Interestingly, no additive or synergistic induction of cytokine production was seen even when gram-positive probiotic bacterial strains were combined with a gram-negative bacteria, E. coli. On the contrary, E. coli-induced IFN-γ production was reduced when different probiotics were present during the stimulation experiments. Gram-positive and gram-negative bacteria have been shown to induce quite different cytokine production patterns.
In human PBMCs gram-positive bacteria induce TNF-α[8–10], IL-12[11–15] and IFN-γ[10111315] while gram-negative bacteria have preferentially been suggested to induce IL-10 production[14]. Gram-positive bacteria or their structural components activate cells via TLR2, whereas gram-negative bacteria and their major structural component lipopolysaccharide activate host cell via TLR4[32]. It is, however, likely that other receptor systems apart from TLRs take part in host cell responses to different microbes[45]. Our data suggests that different bacteria whether they are gram-positive or gram-negative compete with each other during bacteria-host cell interactions. Therefore, combining gram-positive and gram-negative bacteria to activate the respective TLR2 and TLR4-dependent signalling pathways does not further enhance human PBMC responses. Initial analyses with TLR2 and TLR4 ligand combinations also fail to demonstrate any synergistic enhancement in cytokine production (data not shown).
Our results suggest that probiotic bacteria in a genera-specific way direct immune responses to either the Th1 type or the anti-inflammatory side. Besides providing possible explanations for phenomena observed in clinical trials, this finding might enable pinning down the probiotic bacterial (genera) specific factors contributing to the type of immune response elicited. Understanding of the immunological properties of probiotic bacteria is needed in the development of probiotic bacteria for targeted treatment of different disease conditions.
COMMENTS
Background and research frontiers
Probiotic bacteria have been used for the prevention and treatment of a diverse range of disorders. However, the ways in which probiotic bacteria elicit their health effects are not fully understood. One of the action mechanisms could be the ability to induce cytokines that further regulate innate and adaptive immune responses.
Innovations and breakthroughs
At present there is only a limited amount of comparative data available on the ability of different probiotic strains to induce cytokine responses within the same experimental system. Also, the effect of probiotic bacterial combinations on cytokine production in vitro is not well documented although bacterial combinations have been used in many clinical trials. In the present study we have analysed the cytokine production of eleven different potentially probiotic strains from six bacterial genera alone or in combinations in human peripheral blood mononuclear cells in order to indentify potential enhancing or synergistic effects.
Applications
We found that probiotic bacteria direct immune responses to either Th1 type or anti-inflammatory way in a bacterial genera-specific manner. The probiotic S. thermophilus and Leuconostoc strains are more potent inducers of Th1 type cytokines IL-12 and IFN-γ than the probiotic Lactobacillus strains. Bacterial combinations did not result in enhanced cytokine production. More detailed information on the cytokine patterns that probiotic bacteria elicit may help in designing probiotics for specific preventative or therapeutic purposes.
Terminology
Probiotic bacteria are defined as living microorganisms that have beneficial effects on human health.
Peer review
This paper explores a mechanism of action of probiotics, namely the induction of cytokines by different strains of probiotics. The authors have designed a clear, logical study of different strains, dose-response, time of response and interactions of different probiotic strains. The paper includes an excellent exploration of probiotic mixtures.