1. Introduction
Indole-diterpenes are a large structurally diverse group of natural products, many of which are potent tremorgenic mammalian mycotoxins [
1,
2,
3]. This group of metabolites appears to be confined to a limited number of filamentous fungi within the Eurotiomycetes (e.g.,
Penicillium and
Aspergillus spp.) and Sordariomycetes (e.g.,
Epichloë,
Albophoma and
Nodulisporium spp.) [
4]. Indole-diterpenes have a number of biological activities including insect feeding deterrence [
5,
6], modulation of insect and mammalian potassium ion channels [
7,
8], and inhibition of specific enzymes [
9]. These diverse biological activities have made this group of compounds particularly attractive as potentially new bioactive and therapeutic agents.
Using
Penicillium paxilli as a model experimental system we have identified and functionally characterized the genes required for the synthesis of paxilline [
10,
11], a potent inhibitor of calcium activated BK channels [
7]. Genetic analysis of
P. paxilli has established that a cluster of seven genes is required for paxilline biosynthesis [
10,
11,
12]. Using a
P. paxilli mutant deleted for the entire
pax gene cluster we showed by gene reconstitution experiments that just four of these genes,
paxG,
paxM,
paxB, and
paxC, are required for the synthesis of paspaline [
13], the first cyclic indole-diterpene intermediate in this pathway (
Figure 1). Based on this study we proposed a biosynthetic scheme for paspaline biosynthesis [
13]. This scheme has recently been experimentally validated by reconstitution of the pathway in the heterologous host
A. oryzae [
14]. Increased chemical complexity is achieved through enzyme-specific decorations of this core structure through the action of two cytochrome P450 monooxygenases, PaxP, and PaxQ [
12,
15]. These additional steps have also been experimentally validated by reconstitution of paxilline biosynthesis in the heterologous host
A. oryzae [
14].
Figure 1.
Chemical structures of paspaline (a), penitrem A (b), and shearinine A (c).
Figure 1.
Chemical structures of paspaline (a), penitrem A (b), and shearinine A (c).
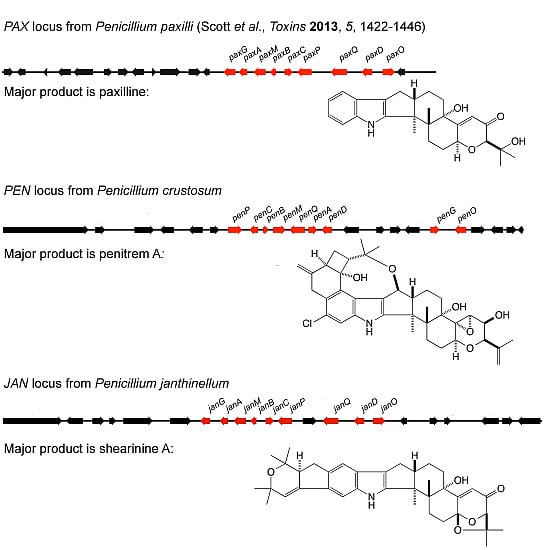
A comparative analysis of related indole-diterpene gene clusters isolated from
Epichloë festucae [
16,
17], a symbiont of forage grasses, and
Aspergillus flavus [
18,
19], has confirmed that these organisms also have the core set of indole-diterpene biosynthetic genes as well as unique genes that are predicted to encode enzyme functions that catalyze the specific chemical decorations that define the predominant indole-diterpene products synthesized by these fungi
i.e., lolitrems (lolitremanes) and aflatrems (aflatremanes), respectively [
20].
Two additional important classes of indole-diterpenes are the penitremanes and janthitremanes [
20], both of which contain compounds that are potent mycotoxins [
21,
22,
23,
24] (
Figure 1). The objective of this study was to clone and analyse gene clusters for indole-diterpene synthesis from strains of
P. crustosum and
P. janthinellum, which synthesize penitremanes and janthitremanes [
25], respectively.
3. Discussion
We describe here the cloning and molecular analysis of two new indole-diterpene gene clusters from
P. crustosum and
P. janthinellum, with genes that encode enzymes for the synthesis of penitrems and shearinines, respectively. Both clusters contain homologs of the core set of four genes,
paxGMBC, shown to be required for paspaline biosynthesis in
P. paxilli [
13], and that are present in all indole-diterpene gene clusters characterized to date [
17,
19], supporting the hypothesis that paspaline is the core cyclic intermediate for the synthesis of a range of indole-diterpenes. In addition, both clusters have homologs of
paxP (
penP/
janP) and
paxQ (
penQ/
janQ), which encode multifunctional cytochrome P450 moonoxygenases for key post-paspaline biosynthetic steps [
15], as well as homologs of
paxA, which encodes an integral membrane protein of unknown function,
paxD, encoding a dimethylallyl transferase [
11], and
paxO, encoding an oxido-reductase that is probably involved in cyclisation of prenylated paxilline.
Both the order and the orientation of these nine genes in the P. janithenllum jan cluster are identical to the corresponding nine pax genes in P. paxilli. However, the one distinct difference between these two clusters is the presence of an extra gene encoding a cytochrome P450 monooxygenase, PJ-13, between janP and janQ. The predicted functions of the genes adjacent to the left- (PJ-05 encoding a ubiquitin ligase and PJ-06 encoding a β-glucosidase) and the right- (PJ-17 encoding a hypothetical protein and PJ-18 encoding an alcohol dehydrogenase) flanks of this cluster would suggest these additional genes have no role in the biosynthesis of janthitremanes. The absence of detectable levels of diprenyl-elaborated indole-diterpenes (including proposed shearinines K and A) in the janP and janD deletion mutants of P. janthinellum and accumulation of the intermediate 13-desoxypaxilline in the latter, provide genetic evidence that this gene cluster is responsible for janthitremane biosynthesis. Our inability to detect paspaline in the janP mutant was surprising but may be associated with the difficulty we have encountered in readily inducing sporulation and secondary metabolite biosynthesis in this particular P. janthinellum strain.
By analogy with the known pathway for paxilline biosynthesis in
P. paxilli [
11,
14], heterologous functional analysis of key biosynthetic steps in aflatrem biosynthesis in
A. flavus [
19,
31], and a comparison of the structures of the main products of these two pathways with the structures of shearinines A and K, a proposed pathway for the synthesis of these compounds in
P. janithinellum is presented in
Figure 6. This scheme proposes that JanG catalyses the synthesis of geranygeranyl diphosphate (GGPP), which condenses with indole 3-glycerol phosphate to form 3-geranylgeranylindole (GG-I) in the presence of JanC, followed by epoxidation and cyclisation steps catalyzed by JanM and JanB, to form paspaline. JanP is proposed to catalyse the conversion of paspapline to 13-desoxypaspaline via β-PC-M6 in a series of α-face oxidations as occurs in both
P. paxilli and
A. flavus [
15,
19,
31].
Like AtmQ from
A. flavus, JanQ is proposed to carry out sequential β-face oxidation steps at C-7 and C-13 of 13-desoxypaspaline to form paspalicine and paspalinine respectively. By analogy to the diprenylation of the indole ring of paxilline by PaxD, JanD is proposed to carry out a similar reaction in
P. janthinellum to form shearinine K [
11,
24,
30,
32]. Further oxidation and cyclisation of this compound by JanO and/or PJ-13 would generate shearinine A [
29,
30].
In contrast to the janthitremanes, the penitremanes are chemically much more complex and would therefore require more enzymes for their synthesis. The gene content and organization of the
P. crustosum pen cluster is correspondingly much more complex than the
P. janthinellum jan cluster. While homologs of the nine genes identified in
P. paxilli and
P. janthinellum were found in this cluster, a number of additional genes encoding enzymes with functions likely to be required for penitremane biosynthesis were identified in this cluster including: PC-05, PC-20, PC-21, and PC-23 encoding cytochrome P450 monooxygenases and PC-22 encoding a dimethylallyl prenyl transferase. Both the order and the orientation of all the genes in this
P. crustosum cluster from PC-05 through to PC-23 (19 genes) are identical to a cluster of genes recently identified in
P. simplicissimum required for penitrem biosynthesis [
33]. The absence of detectable levels of penitrems A, B, D, E, and F, and 13-desoxypaxilline in the
penP deletion mutant of
P. crustosum, but instead an accumulation of paspaline, provides genetic evidence that this gene cluster is responsible for penitrem biosynthesis.
Figure 6.
Proposed biosynthetic scheme for shearinine K and A biosynthesis in P. janthinellum.
Figure 6.
Proposed biosynthetic scheme for shearinine K and A biosynthesis in P. janthinellum.
In a
tour de force Liu
et al. [
33] recently succeeded in elucidating the function of 17 of the proposed 20 genes required for penitrem biosynthesis in
P. simplicissimum by heterologous expression of blocks of these genes in
A. oryzae. On the basis of these reconstitution experiments they propose that PtmGCMB (PenGCMB) are responsible for the synthesis of paspaline and PtmPQ (PenPQ) for subsequent conversion to paxilline, which serves as a substrate for a three step process consisting of C10 ketoreduction (PtmH/PC-15), followed by C20 prenylation (PtmD/PenD) and dehydration (PtmV/PC-16, PtmI/not annotated) to generate the dehydration product of mono-prenylated β-paxitriol. This key intermediate is then converted to PC-M4 via the action of an oxidoreductase (PtmO/PenO) and a second dimethylallyl prenyl transferase (PtmE/PC-22). A series of oxidation steps involving 4 cytochrome P450 monooxygenases (PtmK, PtmU, PtmL, PtmJ/PC-21, PC-05, PC-23, PC-20) and an FAD-dependent monooxygenase (PtmN/PC-14) are required for the transformation of PC-M4 to penitrems A and E. Synthesis of these final products is proposed to proceed via penitrems D and C (PtmK, PtmU, PtmN/PC-21, PC-05, PC-14) and penitrems B and F (PtmK, PtmU, PtmN, PtmL/PC-21, PC-05, PC-14, PC-23). This analysis and earlier work demonstrates the power of the
A. oryzae heterologous gene expression system for biological synthesis of complex natural products [
14,
31,
33].
In summary, the work described here and in earlier papers provides a genetic basis for the diversity of indole-diterpene natural products found within the Penicillium and Aspergillus species. All the indole-diterpene gene clusters identified to date have a core set of genes for the synthesis of paspaline, and a suite of additional genes that encode multi-functional cytochrome P450 monooxygenases, FAD dependent monooxygenases and prenyl transferases that catalyse various regio- and stereo- specific oxidations on this molecular skeleton to generate a diversity of indole-diterpene products.
4. Experimental Section
4.1. Fungal Strains and Growth Conditions
Cultures of wild-type
P. crustosum (isolate PN2402) and
P. janthinellum (isolate PN2408) were maintained on 2.4% Difco potato dextrose agar (Beckton Dickinson, MD, USA) plates or as spore suspensions in 10% (
v/
v) glycerol at −80 °C (
Table A3). For indole-diterpene production, 100 mL Erlenmeyer flasks containing 25 mL of YEPGA medium [
12] were inoculated with 5 × 10
6 spores and grown shaking (200 rpm) for 48 h at 28 °C. Aliquots of 2 mL were used to inoculate 250 mL Erlenmeyer flasks containing 50 mL of aflatrem production medium [
18] which were grown without agitation at 29 °C in the dark for ten days. The mycelial mat that formed on the surface of the liquid was harvested, washed in Milli-Q water (Millipore, MA, USA) and freeze-dried for indole-diterpene analysis. Fungal samples were stored at −80 °C prior to drying or analysis. For isolation of genomic DNA, 25 mL of CDYE medium [
10] was inoculated with 5 × 10
6 spores and grown shaking (200 rpm) for 48 h at 30 °C. Mycelium was harvested, washed in Milli-Q water (Millipore), and freeze dried.
4.2. Isolation, PCR-Amplification, and Sequencing of Genomic DNA
Genomic DNA was isolated from freeze-dried mycelia using the method of Yoder [
34] after grinding in liquid nitrogen with a pestle and mortar. Genomic DNA was amplified using the TripleMaster PCR system (Eppendorf, Hamburg, Germany). Reactions were each performed in a 50 µL volume that contained 1 × High-Fidelity buffer with final concentrations of 4 mM magnesium acetate, 200 µM of each dNTP, 2 U TripleMaster polymerase mixture, 400 nM of each primer, and 50 ng of genomic DNA. Thermal cycling was performed in a Mastercycle gradient thermocycler (Eppendorf) with the following conditions: two min at 94 °C followed by 30 cycles of 94 °C for 30 sec, 65 °C for 30 sec, and 72 °C for 3 min; with a final elongation for 10 min at 72 °C. Primers used for amplification of indole-diterpene cluster sequences are shown in
Table A1. PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) prior to sequencing. At least two independent PCR reactions were combined and sequenced directly on both strands using the dideoxynucleotide chain termination method with the Big-Dye Terminator (version 3.1) Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and separated using an ABI3730 Genetic Analyzer (Applied Biosystems).
4.3. Cosmid Library Production and Screening
Fungal protoplasts were prepared as described previously [
12] except that
P. crustosum cultures were grown shaking for 21 h and
P. janthinellum for 30 h. High molecular weight genomic DNA isolated from protoplasts using the method of Byrd
et al. [
35] was partially digested using
MboI in a reaction containing 7.5 µg gDNA and enzyme concentrations of 0.141 U and 0.126 U per µg gDNA for
P. crustosum and
P. janthinellum, respectively. Reaction volumes of 100 µL were incubated at 37 °C for 60 min. Partially digested gDNA was end-filled with dATP and dGTP to generate 5'-GA protruding termini. Cosmid vector pMOcosX (120 µg) [
36] was digested with 100 U each of
XbaI and
XhoI in a final reaction volume of 300 µL for 2 h at 37 °C to yield left and right cosmid arms. Purified digested cosmid DNA was partially end-filled with dTTP and dCTP to generate protruding 5'-TC termini. End filling reactions were performed in 100 µL reaction volumes containing 4 µg gDNA or 20 µg cosmid DNA with 1 U/µg of the Klenow fragment of DNA polymerase I (Invitrogen, CA, USA) and 0.5 mM of each of dTTP and dCTP (for cosmid DNA) or dATP and dGTP (for gDNA) incubated at room temperature for 20 min. Digestion and end-filling reactions were each stopped and purified by phenol:chloroform extraction according to standard molecular biology techniques [
37] and resuspended in 50 µL MilliQ water.
Ligation reactions containing digested and end-filled cosmid DNA (6 µg) and gDNA (1.5 µg) in final volumes of 60 µL containing 9 Weiss units of T4 DNA ligase (Promega) were incubated at 16 °C for 6 h then stored at 4 °C. Packaging reactions and transduction of host cells were performed using Gigapack III Gold Packaging Extract and
E. coli host strain VCS257, respectively according to the manufacturer’s recommendations (Stratagene, La Jolla, CA, USA). Libraries were amplified and for each, approximately 12,000 ampicillin-resistant transformants were spread on six 90 mm diameter agar plates. The libraries were screened using radioactively labelled hybridization probes according to standard molecular biology techniques [
37]. The
P. crustosum library was screened using a 1448-bp PCR fragment amplified using primers 2402CF1 and 2402PF1 for
P. janthinellum PN2408 a 1045-bp fragment amplified using primers 2408F1 and 2408PF1 (
Table A1) which contained the 3' ends of
paxC and
paxP homologs and corresponding intergenic regions, respectively.
4.4. Cosmid Sequencing
The Illumina Genome Analyzer IIx was used to generate 75-bp paired-end reads, which were processed with version 1.6 of Illumina’s data analysis pipeline. Reads were trimmed to the largest contiguous sequence where quality scores exceeded
P = 0.05 [
38]. After trimming, reads shorter than 25 bp were discarded. Reads were
de novo assembled using the de Bruijn graph assembler
ABySS (version 1.2.0) [
39]. Trimmed and untrimmed reads were assembled separately using a range of
k-mer values [19–61, odd numbers only] and a parameter sweep of
n [
2,
5,
10],
c [
1,
10,
20] and
e [
1,
3,
10]. The assembly with the most complete coverage of the cosmid, as recognized from flanking vector sequence, was extracted manually. Gaps in the assembly were corrected manually by patching with sequence information from other assemblies.
4.6. Preparation of Gene Replacement Constructs and Southern Analysis
The
penP,
janP, and
janD replacement constructs pCE50, pCE51, and pCS5 were prepared by yeast recombination cloning [
41] using
EcoRI/
XhoI cut pRS426, 5' and 3' fragments to each of the genes amplified from genomic DNA using Phusion™ High-Fidelity DNA polymerase (Thermo Scientific, Waltham, MA, USA), together with a 1.38-kb
PtrpC-hph fragment amplified from pSF15.15 using primer pair hph-F/hph-R (
Table A4). The 921-bp 5' and 1271-bp 3' fragments of
penP were amplified using primer pairs pRS426-penP-F/penP-hph-R and hph-penP-F/penP-pRS426-R, respectively. The 1152-bp 5' and 1071-bp 3' fragments of
janP were amplified using primer pairs pRS426-janP-F/janP-hph-R and hph-janP-F/janP-pRS426-R, respectively. The 1205-bp 5' and 1002-bp 3' fragments of
janD were amplified using primer pairs CSPjantD1F/CSPjantD2R and CSPjantD3F/CSPjantD4R, respectively.
To facilitate yeast recombinational cloning, primers for amplification of the penP, janP, and janD 5' flanking fragments contained overlap to the yeast vector pRS426 (pRS426-penP-F, pRS426-janP-F and CSPjantD1F) and to the hph hygromycin resistance cassette (penP-hph-R, janP-hph-R and CSPjantD2R), and primers for amplification of the 3' flanking fragment contained overlap to the hph resistance cassette (hph-penP-F, hph-janP-F and CSPjantD3F) and to pRS426 (penP-pRS426-R, janP-pRS426-R and CSPjantD4R).
Yeast cells were then transformed with
EcoRI/
XhoI linearised pRS426, PCR amplified P
trpC-
hph cassette, and
penP,
janP or
janD 5' and 3' flanking region PCR fragments as previously described [
42]. Transformants were selected on media lacking uracil and plasmid DNA subsequently isolated and transformed into
E. coli. Plasmid DNA was isolated from resulting
E. coli transformants and analysed for the correct construct by diagnostic PCR and DNA sequencing.
Lists of all plasmids and the primer sequences used to prepare those constructs can be found in
Table A3 and
Table A4.
4.8. Indole-Diterpene Chemical Analysis
Freeze dried fungal biomass (0.5 or 1 g) was homogenized in 25 mL of 7:3 (v/v) propan-2-ol-water mixture at ambient temperature for extraction of the indole-diterpenes. After mixing for 1 h, the samples were centrifuged to pellet the insoluble residue. The supernatants were stored at −18 °C.
LC-MS/MS analysis was performed using a system consisting of a Thermo PAL sampler, Thermo Accela pump, and Thermo LTQ XL linear ion trap mass spectrometer in ESI positive ion mode (Thermo Scientific, Waltham, CA, USA). A Luna C18 column (150 × 2 mm, 5 µm); Phenomenex, Torrance, CA, USA) was used for separations at a flow rate of 200 µL/min of a linear gradient over 40 min, beginning at 1:1 acetonitrile-water with 0.1% (v/v) acetic acid through to acetonitrile with 0.1% acetic acid, this held for a further 20 min. The mass spectrometer was setup in data-dependent mode for collection of fragmentation data of a range of preselected ions to assist attribution of identities of indole-diterpene compounds.
4.9. Nucleotide Sequence Accession Numbers
Contiguous sequences containing the PEN cluster from P. crustosum and JAN cluster from P. janthinellum have been deposited with GenBank under accession numbers KC963408 and KF280651, respectively.