Brachypodium Antifreeze Protein Gene Products Inhibit Ice Recrystallisation, Attenuate Ice Nucleation, and Reduce Immune Response
Abstract
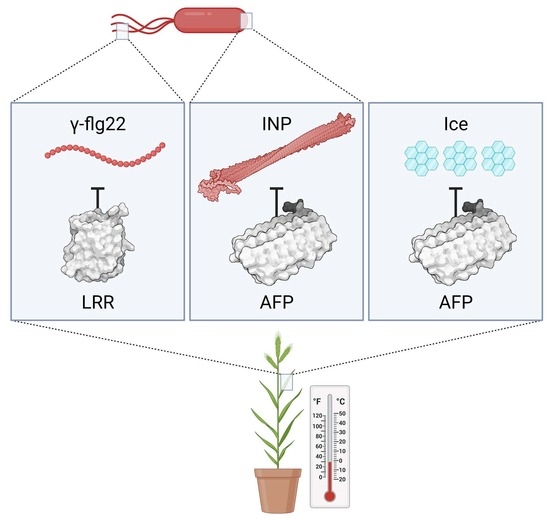
:1. Introduction
2. Results
2.1. AFP-Mediated Attenuation of INP-Induced Freezing and INP Models
2.2. LRR-Mediated Attenuation of the Host Immune Response and Modelling
3. Discussion
4. Materials and Methods
4.1. Protein Modelling and Interaction Predictions
4.2. Construction of LRR Plasmids for Arabidopsis
4.3. Transient Expression in Arabidopsis
4.4. Plant Material and Growth Conditions
4.5. Preparation of Extracts and Apoplast Samples for AFP Activity
4.6. Ice Nucleation Assay
4.7. Immune Response Attenuation ROS Burst Assay
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Copley, S.D. An evolutionary perspective on protein moonlighting. Biochem. Soc. Trans. 2014, 42, 1684–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hon, W.C.; Griffith, M.; Mlynarz, A.; Kwok, Y.C.; Yang, D.S. Antifreeze proteins in winter rye are similar to pathogenesis-related proteins. Plant Physiol. 1995, 109, 879–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.M.; Griffith, M.; Wiseman, S.B. Ethylene induces antifreeze activity in winter rye leaves. Plant Physiol. 2001, 126, 1232–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandve, S.R.; Rudi, H.; Asp, T.; Rognli, O.A. Tracking the evolution of a cold stress associated gene family in cold tolerant grasses. BMC Evol. Biol. 2008, 8, 245. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Rudi, H.; Stockinger, E.J.; Cheng, H.; Cao, M.; Fox, S.E.; Mockler, T.C.; Westereng, B.; Fjellheim, S.; Rognli, O.A.; et al. Comparative analyses reveal potential uses of Brachypodium distachyon as a model for cold stress responses in temperate grasses. BMC Plant Biol. 2012, 12, 65. [Google Scholar] [CrossRef] [Green Version]
- Bredow, M.; Vanderbeld, B.; Walker, V.K. Knockdown of ice-binding proteins in Brachypodium distachyon demonstrates their role in freeze protection. PLoS ONE 2016, 11, e0167941. [Google Scholar] [CrossRef] [Green Version]
- Bredow, M.; Vanderbeld, B.; Walker, V.K. Ice-binding proteins confer freezing tolerance in transgenic Arabidopsis thaliana. Plant Biotechnol. J. 2017, 15, 68–81. [Google Scholar] [CrossRef] [Green Version]
- Juurakko, C.L.; Bredow, M.; diCenzo, G.C.; Walker, V.K. Cold-inducible promoter-driven knockdown of Brachypodium antifreeze proteins confers freeze sensitivity. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bredow, M.; Tomalty, H.E.; Smith, L.; Walker, V.K. Ice and anti-nucleating activities of an ice-binding protein from the annual grass, Brachypodium distachyon. Plant Cell Environ. 2018, 41, 983–992. [Google Scholar] [CrossRef]
- Garnham, C.P.; Campbell, R.L.; Walker, V.K.; Davies, P.L. Novel dimeric β-helical model of an ice nucleation protein with bridged active sites. BMC Struct. Biol. 2011, 11, 36. [Google Scholar] [CrossRef] [Green Version]
- Middleton, A.J.; Marshall, C.B.; Faucher, F.; Bar-Dolev, M.; Braslavsky, I.; Campbell, R.L.; Walker, V.K.; Davies, P.L. Antifreeze protein from freeze-tolerant grass has a beta-roll fold with an irregularly structured ice-binding site. J. Mol. Biol. 2012, 416, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Nemecek-Marshall, M.; Laduca, R.; Fall, R. High-level expression of ice nuclei in a Pseudomonas syringae strain is induced by nutrient limitation and low temperature. J. Bacteriol. 1993, 175, 4062–4070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fall, A.L.; Fall, R. High-level expression of ice nuclei in Erwinia herbicola is induced by phosphate starvation and low temperature. Curr. Microbiol. 1998, 36, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Frauenfeld, O.W.; Cao, B.; Wang, K.; Wang, H.; Su, H.; Huang, Z.; Yue, D.; Zhang, T. Response of changes in seasonal soil freeze/thaw state to climate change from 1950 to 2010 across China. J. Geophys. Res. Earth Surf. 2016, 121, 1984–2000. [Google Scholar] [CrossRef]
- Smith, A.; Lott, N.; Houston, T.; Shein, K.; Crouch, J.; Enloe, J. US Billion-Dollar Weather Climate Disasters 1980–2021; NOAA National Centers for Environmental Information: Asheville, NC, USA, 2020; p. 15. [Google Scholar]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nürnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Cao, Y.; Aceti, D.J.; Sabat, G.; Song, J.; Makino, S.I.; Fox, B.G.; Bent, A.F. Mutations in FLS2 Ser-938 dissect signaling activation in FLS2-mediated Arabidopsis immunity. PLoS Pathog. 2013, 9, e1003313. [Google Scholar] [CrossRef] [Green Version]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Garzón, J.I.; Lopéz-Blanco, J.R.; Pons, C.; Kovacs, J.; Abagyan, R.; Fernández-Recio, J.; Chacón, P. FRODOCK: A new approach for fast rotational protein–protein docking. Bioinformatics 2009, 25, 2544–2551. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Aportela, E.; López-Blanco, J.R.; Chacón, P. FRODOCK 2.0: Fast protein–protein docking server. Bioinformatics 2016, 32, 2386–2388. [Google Scholar] [CrossRef] [Green Version]
- Hudait, A.; Odendahl, N.; Qiu, Y.; Paesani, F.; Molinero, V. Ice-nucleating and antifreeze proteins recognize ice through a diversity of anchored clathrate and ice-like motifs. J. Am. Chem. Soc. 2018, 140, 4905–4912. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.L.; Wex, H.; Grawe, S.; Jakobsson, J.; Löndahl, J.; Hartmann, S.; Finster, K.; Boesen, T.; Šantl-Temkiv, T. Effects of ice nucleation protein repeat number and oligomerization level on ice nucleation activity. J. Geophys. Res. Atmos. 2018, 123, 1802–1810. [Google Scholar] [CrossRef]
- Squire, J.M.; Parry, D.A.; Kajava, A. Fibrous proteins: Amyloids, prions and beta proteins. In Advances in Protein Chemistry II; Elsevier: San Diego, CA, USA, 2006; pp. 1–13. [Google Scholar]
- Bryan, A.W.; Starner-Kreinbrink, J.L.; Hosur, R.; Clark, P.L.; Berger, B. Structure-based prediction reveals capping motifs that inhibit β-helix aggregation. Proc. Natl. Acad. Sci. USA 2011, 108, 11099–11104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peralta, M.D.; Karsai, A.; Ngo, A.; Sierra, C.; Fong, K.T.; Hayre, N.R.; Mirzaee, N.; Ravikumar, K.M.; Kluber, A.J.; Chen, X.; et al. Engineering amyloid fibrils from β-solenoid proteins for biomaterials applications. ACS Nano 2015, 9, 449–463. [Google Scholar] [CrossRef]
- Middleton, A.J.; Brown, A.M.; Davies, P.L.; Walker, V.K. Identification of the ice-binding face of a plant antifreeze protein. FEBS Lett. 2009, 583, 815–819. [Google Scholar] [CrossRef] [Green Version]
- Tomalty, H.E.; Walker, V.K. Perturbation of bacterial ice nucleation activity by a grass antifreeze protein. Biochem. Biophys. Res. Commun. 2014, 452, 636–641. [Google Scholar] [CrossRef]
- Schwidetzky, R.; Kunert, A.T.; Bonn, M.; Pöschl, U.; Ramløv, H.; DeVries, A.L.; Fröhlich-Nowoisky, J.; Meister, K. Inhibition of bacterial ice nucleators is not an intrinsic property of antifreeze proteins. J. Phys. Chem. B 2020, 124, 4889–4895. [Google Scholar] [CrossRef]
- Roeters, S.J.; Golbek, T.W.; Bregnhøj, M.; Drace, T.; Alamdari, S.; Roseboom, W.; Kramer, G.; Šantl-Temkiv, T.; Finster, K.; Pfaendtner, J.; et al. Ice-nucleating proteins are activated by low temperatures to control the structure of interfacial water. Nat. Commun. 2021, 12, 1183. [Google Scholar] [CrossRef]
- Ono, K.; Hasegawa, K.; Naiki, H.; Yamada, M. Anti-amyloidogenic activity of tannic acid and its activity to destabilize Alzheimer’s β-amyloid fibrils in vitro. Biochim. Biophys. Acta Mol. Basis Dis. 2004, 1690, 193–202. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Hudait, A.; Molinero, V. How size and aggregation of ice-binding proteins control their ice nucleation efficiency. J. Am. Chem. Soc. 2019, 141, 7439–7452. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.H.; Bredow, M.; Monaghan, J.; diCenzo, G.C. Proteobacteria contain diverse flg22 epitopes that elicit varying immune responses in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2021, 34, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.M.; Chai, J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013, 342, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Tsaban, T.; Varga, J.K.; Avraham, O.; Ben-Aharon, Z.; Khramushin, A.; Schueler-Furman, O. Harnessing protein folding neural networks for peptide–protein docking. Nat. Commun. 2022, 13, 1–12. [Google Scholar] [CrossRef]
- Juurakko, C.L.; diCenzo, G.C.; Walker, V.K. Cold acclimation and prospects for cold-resilient crops. Plant Stress 2021, 2, 100028. [Google Scholar] [CrossRef]
- Bostock, R.M.; Pye, M.F.; Roubtsova, T.V. Predisposition in plant disease: Exploiting the nexus in abiotic and biotic stress perception and response. Annu. Rev. Phytopathol. 2014, 52, 517–549. [Google Scholar] [CrossRef] [Green Version]
- Saijo, Y.; Loo, E.P. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef] [Green Version]
- Govindarajan, A.G.; Lindow, S.E. Size of bacterial ice-nucleation sites measured in situ by radiation inactivation analysis. Proc. Natl. Acad. Sci. USA 1988, 85, 1334–1338. [Google Scholar] [CrossRef] [Green Version]
- Lukas, M.; Schwidetzky, R.; Kunert, A.T.; Poschl, U.; Frohlich-Nowoisky, J.; Bonn, M.; Meister, K. Electrostatic interactions control the functionality of bacterial ice nucleators. J. Am. Chem. Soc. 2020, 142, 6842–6846. [Google Scholar] [CrossRef]
- Fletcher, G.L.; Hew, C.L.; Davies, P.L. Antifreeze proteins of teleost fishes. Annu. Rev. Physiol. 2001, 63, 359–390. [Google Scholar] [CrossRef] [Green Version]
- Ali, G.S.; Reddy, A.S.N. PAMP-triggered immunity: Early events in the activation of FLAGELLIN SENSITIVE2. Plant Signal. Behav. 2008, 3, 423–426. [Google Scholar] [CrossRef]
- Kimura, S.; Waszczak, C.; Hunter, K.; Wrzaczek, M. Bound by fate: The role of reactive oxygen species in receptor-like kinase signaling. Plant Cell 2017, 29, 638–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rämisch, S.; Weininger, U.; Martinsson, J.; Akke, M.; André, I. Computational design of a leucine-rich repeat protein with a predefined geometry. Proc. Natl. Acad. Sci. USA 2014, 111, 17875–17880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juurakko, C.L.; diCenzo, G.C.; Walker, V.K. Cold acclimation in Brachypodium is accompanied by changes in above-ground bacterial and fungal communities. Plants 2021, 10, 2824. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 2022, 1–3. [Google Scholar] [CrossRef]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022, 13, 1265. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.W.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Tang, J.; Han, Z.; Sun, Y.; Zhang, H.; Gong, X.; Chai, J. Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 2015, 25, 110–120. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krissinel, E.; Henrick, K. Detection of protein assemblies in crystals. In International Symposium on Computational Life Science; Springer: Berlin/Heidelberg, Germany, 2005; pp. 163–174. [Google Scholar]
- Krissinel, E.; Henrick, K. Protein interfaces, surfaces and assemblies service PISA at European Bioinformatics Institute. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Krissinel, E. Crystal contacts as nature’s docking solutions. J. Comput. Chem. 2010, 31, 133–143. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, M.; Siemiatkowska, B.; Toleco, M.R.; Jing, Y.; Strotmann, V.; Zhang, J.; Stahl, Y.; Fernie, A.R. A highly efficient agrobacterium-mediated method for transient gene expression and functional studies in multiple plant species. Plant Commun. 2020, 1, 100028. [Google Scholar] [CrossRef] [PubMed]
- Bredow, M.; Tomalty, H.E.; Graham, L.A.; Gruneberg, A.K.; Middleton, A.J.; Vanderbeld, B.; Davies, P.L.; Walker, V.K. Isolation and characterization of ice-binding proteins from higher plants. In Plant Cold Acclimation; Humana Press: New York, NY, USA; pp. 303–332.
- Pogorelko, G.; Fursova, O.; Lin, M.; Pyle, E.; Jass, J.; Zabotina, O.A. Post-synthetic modification of plant cell walls by expression of microbial hydrolases in the apoplast. Plant Mol. Biol. 2011, 77, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Vali, G. Quantitative evaluation of experimental results on heterogeneous freezing nucleation of supercooled liquids. J. Atmos. Sci. 1971, 28, 402–409. [Google Scholar] [CrossRef]
- Trujillo, M. Analysis of the immunity-related oxidative bursts by a luminol-based assay. In Environmental Responses in Plants; Humana Press: New York, NY, USA; pp. 323–329.






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juurakko, C.L.; diCenzo, G.C.; Walker, V.K. Brachypodium Antifreeze Protein Gene Products Inhibit Ice Recrystallisation, Attenuate Ice Nucleation, and Reduce Immune Response. Plants 2022, 11, 1475. https://doi.org/10.3390/plants11111475
Juurakko CL, diCenzo GC, Walker VK. Brachypodium Antifreeze Protein Gene Products Inhibit Ice Recrystallisation, Attenuate Ice Nucleation, and Reduce Immune Response. Plants. 2022; 11(11):1475. https://doi.org/10.3390/plants11111475
Chicago/Turabian StyleJuurakko, Collin L., George C. diCenzo, and Virginia K. Walker. 2022. "Brachypodium Antifreeze Protein Gene Products Inhibit Ice Recrystallisation, Attenuate Ice Nucleation, and Reduce Immune Response" Plants 11, no. 11: 1475. https://doi.org/10.3390/plants11111475