The Epigenetic Legacy of Maternal Protein Restriction: Renal Ptger1 DNA Methylation Changes in Hypertensive Rat Offspring
Abstract
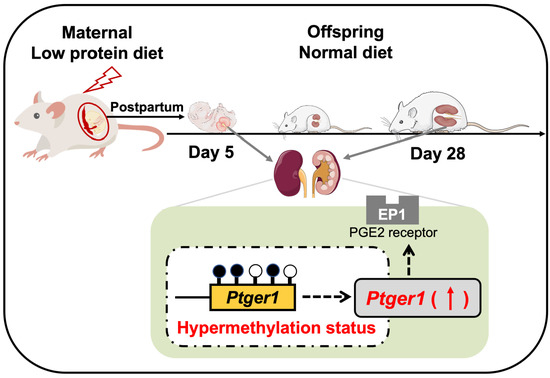
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.1.1. Prenatal Low-Protein Diet
2.1.2. Postnatal Low and High-Protein Diets
2.2. DNA Extraction and Methylome Analysis
2.3. Bisulfite Sequencing
2.4. RNA Extraction and qPCR
2.5. DNA Microarray Analysis
2.6. Statistical Analysis
3. Results
3.1. Identification of Candidate Genes for Salt-Sensitive Hypertension Using D28 Pups Exposed to a Maternal Low-Protein Diet
3.2. DNA Methylation Status of Ptger1 CpG Island in D28 Pup Kidneys
3.3. DNA Methylation Status of the Ptger1 Promoter Region in D28 Pup Kidneys
3.4. Ptger1 mRNA Expression from D5 to D28 in the Kidneys of Pups Exposed to a Fetal Low-Protein Diet
3.5. DNA Methylation Status of Ptger1 CpG Islands from D5 to D28 in the Kidneys of Pups Exposed to a Fetal Low-Protein Diet
3.6. Ptger1 mRNA Expression in the Kidneys of D42 Pups Exposed to Postnatal Low- and High-Protein Diets
3.7. DNA Methylation Status of Ptger1 CpG Island in the D42 Pup Kidneys Exposed to Postnatal Low- and High-Protein Diets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marshall, N.E.; Abrams, B.; Barbour, L.A.; Catalano, P.; Christian, P.; Friedman, J.E.; Hay, W.W., Jr.; Hernandez, T.L.; Krebs, N.F.; Oken, E.; et al. The importance of nutrition in pregnancy and lactation: Lifelong consequences. Am. J. Obstet. Gynecol. 2022, 226, 607–632. [Google Scholar] [CrossRef] [PubMed]
- Di Costanzo, M.; De Paulis, N.; Capra, M.E.; Biasucci, G. Nutrition during Pregnancy and Lactation: Epigenetic Effects on Infants’ Immune System in Food Allergy. Nutrients 2022, 14, 1766. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.N.; Tain, Y.L. The Good, the Bad, and the Ugly of Pregnancy Nutrients and Developmental Programming of Adult Disease. Nutrients 2019, 11, 894. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.J.; Meaney, M.J. Fetal Origins of Mental Health: The Developmental Origins of Health and Disease Hypothesis. Am. J. Psychiatry 2017, 174, 319–328. [Google Scholar] [CrossRef]
- Hagemann, E.; Silva, D.T.; Davis, J.A.; Gibson, L.Y.; Prescott, S.L. Developmental Origins of Health and Disease (DOHaD): The importance of life-course and transgenerational approaches. Paediatr. Respir. Rev. 2021, 40, 3–9. [Google Scholar] [CrossRef]
- Hoffman, D.J.; Powell, T.L.; Barrett, E.S.; Hardy, D.B. Developmental origins of metabolic diseases. Physiol. Rev. 2021, 101, 739–795. [Google Scholar] [CrossRef]
- Hanson, M.A.; Gluckman, P.D. Early developmental conditioning of later health and disease: Physiology or pathophysiology? Physiol. Rev. 2014, 94, 1027–1076. [Google Scholar] [CrossRef]
- Wu, T.; Zhou, K.; Hua, Y.; Zhang, W.; Li, Y. The molecular mechanisms in prenatal drug exposure-induced fetal programmed adult cardiovascular disease. Front. Pharmacol. 2023, 14, 1164487. [Google Scholar] [CrossRef]
- Fall, C.H.D.; Kumaran, K. Metabolic programming in early life in humans. Philos. Trans. R. Soc. B 2019, 374, 20180123. [Google Scholar] [CrossRef]
- Hsu, C.N.; Tain, Y.L. The Double-Edged Sword Effects of Maternal Nutrition in the Developmental Programming of Hypertension. Nutrients 2018, 10, 1917. [Google Scholar] [CrossRef]
- Paauw, N.D.; van Rijn, B.B.; Lely, A.T.; Joles, J.A. Pregnancy as a critical window for blood pressure regulation in mother and child: Programming and reprogramming. Acta Physiol. 2017, 219, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Langley-Evans, S.C. Nutritional programming of disease: Unravelling the mechanism. J. Anat. 2009, 215, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Nusken, E.; Dotsch, J.; Weber, L.T.; Nusken, K.D. Developmental Programming of Renal Function and Re-Programming Approaches. Front. Pediatr. 2018, 6, 36. [Google Scholar] [CrossRef]
- Whitehouse, A.J.; Holt, B.J.; Serralha, M.; Holt, P.G.; Kusel, M.M.; Hart, P.H. Maternal serum vitamin D levels during pregnancy and offspring neurocognitive development. Pediatrics 2012, 129, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Mathias, P.C.; Elmhiri, G.; de Oliveira, J.C.; Delayre-Orthez, C.; Barella, L.F.; Tofolo, L.P.; Fabricio, G.S.; Chango, A.; Abdennebi-Najar, L. Maternal diet, bioactive molecules, and exercising as reprogramming tools of metabolic programming. Eur. J. Nutr. 2014, 53, 711–722. [Google Scholar] [CrossRef]
- Eitmann, S.; Matrai, P.; Nemeth, D.; Hegyi, P.; Lukacs, A.; Berczi, B.; Czumbel, L.M.; Kiss, I.; Gyongyi, Z.; Varga, G.; et al. Maternal overnutrition elevates offspring’s blood pressure-A systematic review and meta-analysis. Paediatr. Perinat. Epidemiol. 2022, 36, 276–287. [Google Scholar] [CrossRef]
- Odhiambo, J.F.; Pankey, C.L.; Ghnenis, A.B.; Ford, S.P. A Review of Maternal Nutrition during Pregnancy and Impact on the Offspring through Development: Evidence from Animal Models of Over- and Undernutrition. Int. J. Environ. Res. Public Health 2020, 17, 6926. [Google Scholar] [CrossRef]
- Vickers, M.H. Utility of preclinical models of altered maternal nutrition to support the developmental origins of health and disease hypothesis. Clin. Sci. 2022, 136, 711–714. [Google Scholar] [CrossRef]
- Hult, M.; Tornhammar, P.; Ueda, P.; Chima, C.; Bonamy, A.K.; Ozumba, B.; Norman, M. Hypertension, diabetes and overweight: Looming legacies of the Biafran famine. PLoS ONE 2010, 5, e13582. [Google Scholar] [CrossRef]
- Stein, A.D.; Zybert, P.A.; van der Pal-de Bruin, K.; Lumey, L.H. Exposure to famine during gestation, size at birth, and blood pressure at age 59 y: Evidence from the Dutch Famine. Eur. J. Epidemiol. 2006, 21, 759–765. [Google Scholar] [CrossRef]
- Hrudey, E.J.; Reynolds, R.M.; Oostvogels, A.J.; Brouwer, I.A.; Vrijkotte, T.G. The Association between Maternal 25-Hydroxyvitamin D Concentration during Gestation and Early Childhood Cardio-metabolic Outcomes: Is There Interaction with Pre-Pregnancy BMI? PLoS ONE 2015, 10, e0133313. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.M.; Fraser, A.; Fraser, W.D.; Hypponen, E.; Davey Smith, G.; Deanfield, J.; Hingorani, A.; Sattar, N.; Lawlor, D.A. Associations of maternal 25-hydroxyvitamin D in pregnancy with offspring cardiovascular risk factors in childhood and adolescence: Findings from the Avon Longitudinal Study of Parents and Children. Heart 2013, 99, 1849–1856. [Google Scholar] [CrossRef] [PubMed]
- Bergel, E.; Belizan, J.M. A deficient maternal calcium intake during pregnancy increases blood pressure of the offspring in adult rats. BJOG 2002, 109, 540–545. [Google Scholar] [CrossRef]
- Gambling, L.; Dunford, S.; Wallace, D.I.; Zuur, G.; Solanky, N.; Srai, S.K.; McArdle, H.J. Iron deficiency during pregnancy affects postnatal blood pressure in the rat. J. Physiol. 2003, 552, 603–610. [Google Scholar] [CrossRef]
- Tomat, A.; Elesgaray, R.; Zago, V.; Fasoli, H.; Fellet, A.; Balaszczuk, A.M.; Schreier, L.; Costa, M.A.; Arranz, C. Exposure to zinc deficiency in fetal and postnatal life determines nitric oxide system activity and arterial blood pressure levels in adult rats. Br. J. Nutr. 2010, 104, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Selvaraju, V.; Babu, J.R.; Geetha, T. Association of the DNA Methylation of Obesity-Related Genes with the Dietary Nutrient Intake in Children. Nutrients 2023, 15, 2840. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.J.; Vrana, P.B.; Rosenfeld, C.S. Maternal methyl supplemented diets and effects on offspring health. Front. Genet. 2014, 5, 289. [Google Scholar] [CrossRef]
- McGee, M.; Bainbridge, S.; Fontaine-Bisson, B. A crucial role for maternal dietary methyl donor intake in epigenetic programming and fetal growth outcomes. Nutr. Rev. 2018, 76, 469–478. [Google Scholar] [CrossRef]
- Lerman, L.O.; Kurtz, T.W.; Touyz, R.M.; Ellison, D.H.; Chade, A.R.; Crowley, S.D.; Mattson, D.L.; Mullins, J.J.; Osborn, J.; Eirin, A.; et al. Animal Models of Hypertension: A Scientific Statement From the American Heart Association. Hypertension 2019, 73, e87–e120. [Google Scholar] [CrossRef]
- Hsu, C.N.; Tain, Y.L. Animal Models for DOHaD Research: Focus on Hypertension of Developmental Origins. Biomedicines 2021, 9, 623. [Google Scholar] [CrossRef]
- Kato, T.; Mizuguchi, N.; Ito, A. Blood pressure, renal biochemical parameters and histopathology in an original rat model of essential hypertension (SHRSP/Kpo strain). Biomed. Res. 2015, 36, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Aragane, Y.; Higashino, T.; Kinoshita, K.; Ashenagar, M.S.; Higashino, H. Hypertension-Associated Genes in the Mesenteric Artery of Three Spontaneously Hypertensive Rat Substrains Identified Using a DNA Array Method. Front. Biosci. 2022, 27, 191. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, F.; Liang, Y.Q.; Isono, M.; Tajima, M.; Cui, Z.H.; Iizuka, Y.; Gotoda, T.; Nabika, T.; Kato, N. Integrative genomic analysis of blood pressure and related phenotypes in rats. Dis. Models Mech. 2021, 14, 8090. [Google Scholar] [CrossRef]
- Skvortsova, K.; Iovino, N.; Bogdanovic, O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018, 19, 774–790. [Google Scholar] [CrossRef]
- Breton, C.V.; Landon, R.; Kahn, L.G.; Enlow, M.B.; Peterson, A.K.; Bastain, T.; Braun, J.; Comstock, S.S.; Duarte, C.S.; Hipwell, A.; et al. Exploring the evidence for epigenetic regulation of environmental influences on child health across generations. Commun. Biol. 2021, 4, 769. [Google Scholar] [CrossRef] [PubMed]
- Siddeek, B.; Simeoni, U. Epigenetics provides a bridge between early nutrition and long-term health and a target for disease prevention. Acta Paediatr. 2022, 111, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Care, A.S.; Sung, M.M.; Panahi, S.; Gragasin, F.S.; Dyck, J.R.; Davidge, S.T.; Bourque, S.L. Perinatal Resveratrol Supplementation to Spontaneously Hypertensive Rat Dams Mitigates the Development of Hypertension in Adult Offspring. Hypertension 2016, 67, 1038–1044. [Google Scholar] [CrossRef]
- Franzago, M.; Santurbano, D.; Vitacolonna, E.; Stuppia, L. Genes and Diet in the Prevention of Chronic Diseases in Future Generations. Int. J. Mol. Sci. 2020, 21, 2633. [Google Scholar] [CrossRef]
- Otani, L.; Shirasaka, N.; Yoshizumi, H.; Murakami, T. The effects of maternal mild protein restriction on stroke incidence and blood pressure in stroke-prone spontaneously hypertensive rats (SHRSP). Biosci. Biotechnol. Biochem. 2004, 68, 488–494. [Google Scholar] [CrossRef]
- Altobelli, G.; Bogdarina, I.G.; Stupka, E.; Clark, A.J.; Langley-Evans, S. Genome-wide methylation and gene expression changes in newborn rats following maternal protein restriction and reversal by folic acid. PLoS ONE 2013, 8, e82989. [Google Scholar] [CrossRef]
- Paternain, L.; Martisova, E.; Campion, J.; Martinez, J.A.; Ramirez, M.J.; Milagro, F.I. Methyl donor supplementation in rats reverses the deleterious effect of maternal separation on depression-like behaviour. Behav. Brain Res. 2016, 299, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Nakazaki, M.; Oka, S.; Sasaki, M.; Kataoka-Sasaki, Y.; Nagahama, H.; Hashi, K.; Kocsis, J.D.; Honmou, O. Prolonged lifespan in a spontaneously hypertensive rat (stroke prone) model following intravenous infusion of mesenchymal stem cells. Heliyon 2020, 6, e05833. [Google Scholar] [CrossRef] [PubMed]
- Hirao, A.; Tahara, Y.; Kimura, I.; Shibata, S. A balanced diet is necessary for proper entrainment signals of the mouse liver clock. PLoS ONE 2009, 4, e6909. [Google Scholar] [CrossRef] [PubMed]
- Miura, F.; Shibata, Y.; Miura, M.; Sangatsuda, Y.; Hisano, O.; Araki, H.; Ito, T. Highly efficient single-stranded DNA ligation technique improves low-input whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2019, 47, e85. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, M.; Sato, M.; Saito, K.; Otani, L.; Shirahige, K.; Miura, F.; Ito, T.; Jia, H.; Kato, H. Maternal Protein Restriction Alters the Renal Ptger1 DNA Methylation State in SHRSP Offspring. Nutrients 2018, 10, 1436. [Google Scholar] [CrossRef] [PubMed]
- Kamihara, T.; Hirashiki, A.; Kokubo, M.; Shimizu, A. Transcriptome Discovery of Genes in the Three Phases of Autophagy That Are Upregulated During Atrial Fibrillation. Circ. Rep. 2023, 5, 114–122. [Google Scholar] [CrossRef]
- The International Consortium for Blood Pressure Genome-Wide Association Studies; Ehret, G.B.; Munroe, P.B.; Rice, K.M.; Bochud, M.; Johnson, A.D.; Chasman, D.I.; Smith, A.V.; Tobin, M.D.; Verwoert, G.C.; et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011, 478, 103–109. [Google Scholar] [CrossRef]
- Bartlett, C.S.; Boyd, K.L.; Harris, R.C.; Zent, R.; Breyer, R.M. EP1 disruption attenuates end-organ damage in a mouse model of hypertension. Hypertension 2012, 60, 1184–1191. [Google Scholar] [CrossRef]
- Jeong, E.M.; Pereira, M.; So, E.Y.; Wu, K.Q.; Del Tatto, M.; Wen, S.; Dooner, M.S.; Dubielecka, P.M.; Reginato, A.M.; Ventetuolo, C.E.; et al. Targeting RUNX1 as a novel treatment modality for pulmonary arterial hypertension. Cardiovasc. Res. 2022, 118, 3211–3224. [Google Scholar] [CrossRef]
- Wensing, L.A.; Campos, A.H. TBX3, a downstream target of TGF-beta1, inhibits mesangial cell apoptosis. Exp. Cell Res. 2014, 328, 340–350. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, H.; Wang, X.; Snieder, H.; Huang, Y.; Harshfield, G.A.; Treiber, F.A.; Dong, Y. Effects of dopamine receptor type 1 and Gs protein alpha subunit gene polymorphisms on blood pressure at rest and in response to stress. Am. J. Hypertens. 2006, 19, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Wieneke, H.; Svendsen, J.H.; Lande, J.; Spencker, S.; Martinez, J.G.; Strohmer, B.; Toivonen, L.; Le Marec, H.; Garcia-Fernandez, F.J.; Corrado, D.; et al. Polymorphisms in the GNAS Gene as Predictors of Ventricular Tachyarrhythmias and Sudden Cardiac Death: Results From the DISCOVERY Trial and Oregon Sudden Unexpected Death Study. J. Am. Heart Assoc. 2016, 5, e003905. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Fang, S.; Lu, K.T.; Wackman, K.; Schwartzman, M.L.; Dikalov, S.I.; Grobe, J.L.; Sigmund, C.D. EP3 (E-Prostanoid 3) Receptor Mediates Impaired Vasodilation in a Mouse Model of Salt-Sensitive Hypertension. Hypertension 2021, 77, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Saleh, L.S.; Vanderheyden, C.; Frederickson, A.; Bryant, S.J. Prostaglandin E2 and Its Receptor EP2 Modulate Macrophage Activation and Fusion in Vitro. ACS Biomater. Sci. Eng. 2020, 6, 2668–2681. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Fang, B.; Du, S.; Wang, S.; Li, Q.; Jia, X.; Bao, C.; Ye, L.; Sui, X.; Qian, L.; et al. Endothelial cell prostaglandin E2 receptor EP4 is essential for blood pressure homeostasis. JCI Insight 2020, 5, e138505. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, R.; Zimpelmann, J.; Eckert, D.; Ghossein, J.; Geddes, S.; Beique, J.C.; Thibodeau, J.F.; Kennedy, C.R.J.; Burns, K.D.; Hebert, R.L. PGE2 EP1 receptor inhibits vasopressin-dependent water reabsorption and sodium transport in mouse collecting duct. Lab. Investig. 2018, 98, 360–370. [Google Scholar] [CrossRef]
- Makino, H.; Tanaka, I.; Mukoyama, M.; Sugawara, A.; Mori, K.; Muro, S.; Suganami, T.; Yahata, K.; Ishibashi, R.; Ohuchida, S.; et al. Prevention of diabetic nephropathy in rats by prostaglandin E receptor EP1-selective antagonist. J. Am. Soc. Nephrol. 2002, 13, 1757–1765. [Google Scholar] [CrossRef]
- Lam, L.L.; Emberly, E.; Fraser, H.B.; Neumann, S.M.; Chen, E.; Miller, G.E.; Kobor, M.S. Factors underlying variable DNA methylation in a human community cohort. Proc. Natl. Acad. Sci. USA 2012, 109 (Suppl. S2), 17253–17260. [Google Scholar] [CrossRef]
- Feiner, L.K.; Tierling, S.; Hollander, S.; Glanemann, M.; Rubie, C. An aging and p53 related marker: HOXA5 promoter methylation negatively correlates with mRNA and protein expression in old age. Aging 2021, 13, 4831–4849. [Google Scholar] [CrossRef]
- DuBois, B.; Pearson, J.; Hastings, B.; Mahmood, T.; Chan, T.; Alnakhli, A.; Cherala, G. Maternal low-protein diet alters the expression of real-time quantitative polymerase chain reaction reference genes in an age-, sex-, and organ-dependent manner in rat offspring. Nutr. Res. 2013, 33, 235–241. [Google Scholar] [CrossRef]
- Gluckman, P.D.; Hanson, M.A.; Cooper, C.; Thornburg, K.L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008, 359, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J.B.; Yzydorczyk, C.; Siddeek, B.; Peyter, A.C.; Simeoni, U. Intrauterine growth restriction: Clinical consequences on health and disease at adulthood. Reprod. Toxicol. 2021, 99, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Lukaszewski, M.A.; Mayeur, S.; Fajardy, I.; Delahaye, F.; Dutriez-Casteloot, I.; Montel, V.; Dickes-Coopman, A.; Laborie, C.; Lesage, J.; Vieau, D.; et al. Maternal prenatal undernutrition programs adipose tissue gene expression in adult male rat offspring under high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E548–E559. [Google Scholar] [CrossRef] [PubMed]
- Parlee, S.D.; MacDougald, O.A. Maternal nutrition and risk of obesity in offspring: The Trojan horse of developmental plasticity. Biochim. Biophys. Acta 2014, 1842, 495–506. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, H.; Miyoshi, M.; Li, X.; Furukawa, K.; Otani, L.; Shirahige, K.; Miura, F.; Ito, T.; Kato, H. The Epigenetic Legacy of Maternal Protein Restriction: Renal Ptger1 DNA Methylation Changes in Hypertensive Rat Offspring. Nutrients 2023, 15, 3957. https://doi.org/10.3390/nu15183957
Jia H, Miyoshi M, Li X, Furukawa K, Otani L, Shirahige K, Miura F, Ito T, Kato H. The Epigenetic Legacy of Maternal Protein Restriction: Renal Ptger1 DNA Methylation Changes in Hypertensive Rat Offspring. Nutrients. 2023; 15(18):3957. https://doi.org/10.3390/nu15183957
Chicago/Turabian StyleJia, Huijuan, Moe Miyoshi, Xuguang Li, Kyohei Furukawa, Lila Otani, Katsuhiko Shirahige, Fumihito Miura, Takashi Ito, and Hisanori Kato. 2023. "The Epigenetic Legacy of Maternal Protein Restriction: Renal Ptger1 DNA Methylation Changes in Hypertensive Rat Offspring" Nutrients 15, no. 18: 3957. https://doi.org/10.3390/nu15183957