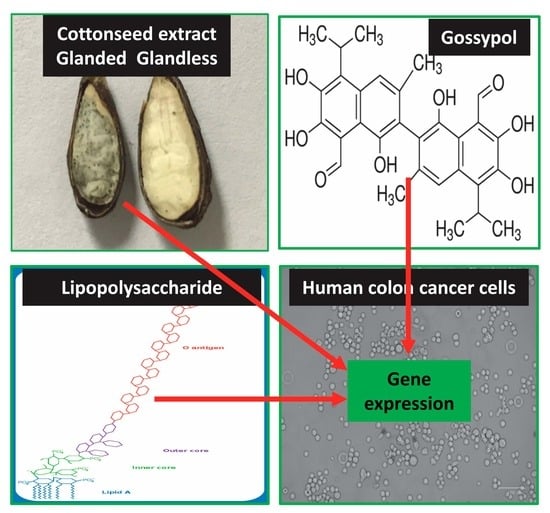
Identification of Bcl2 as a Stably Expressed qPCR Reference Gene for Human Colon Cancer Cells Treated with Cottonseed-Derived Gossypol and Bioactive Extracts and Bacteria-Derived Lipopolysaccharides
Abstract
:1. Introduction
2. Results
2.1. Cq mean Distribution in Colon Cancer Cells Treated with Gossypol, LPS, and Cottonseed Extracts
2.2. Cq standard Deviation Distribution in Colon Cancer Cells Treated with Gossypol, LPS, and Cottonseed Extracts
2.3. Variation of Gene Expression between Plant Toxin Gossypol and Bacterial Toxin LPS Treatment
2.4. Variation of Gene Expression between Glanded and Glandless Cottonseed Extract Treatment
2.5. Variation of Gene Expression between Cottonseed Coat and Kernel Extract Treatment
2.6. Variation of Gene Expression between Cottonseed Extract Treatment Time
2.7. Variation of Gene Expression between DMSO Control and Various Treatments
3. Discussion
4. Materials and Methods
4.1. Colon Cancer Cell Line
4.2. Gossypol, LPS, and Cottonseed Extracts
4.3. Reagents and Equipment
4.4. Cell Culture and Chemical Treatment
4.5. Real-Time qPCR Primers and Probes
4.6. RNA Isolation and cDNA Synthesis
4.7. Quantitative Real-Time PCR Analysis
4.8. Data Analysis and Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cao, H.; Sethumadhavan, K.; Bland, J.M. Isolation of cottonseed extracts that affect human cancer cell growth. Sci. Rep. 2018, 8, 10458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pons, W.A.; Hoffpauir, C.L.; Hopper, T.H. Gossypol in Cottonseed, Influence of Variety of Cottonseed and Environment. J. Agric. Food Chem. 1953, 1, 1115–1118. [Google Scholar] [CrossRef]
- Cao, H.; Sethumadhavan, K. Cottonseed bioactive compounds and peptides. In Proceedings of the 2020 Beltwide Cotton Conferences, Austin, TX, USA, 8–10 January 2020; National Cotton Council of America: Cordova, TN, USA; pp. 270–281. [Google Scholar]
- Piccinelli, A.L.; Veneziano, A.; Passi, S.; Simone, F.D.; Rastrelli, L. Flavonol glycosides from whole cottonseed by-product. Food Chem. 2007, 100, 344–349. [Google Scholar] [CrossRef]
- Wang, X.; Howell, C.P.; Chen, F.; Yin, J.; Jiang, Y. Gossypol—A polyphenolic compound from cotton plant. Adv. Food Nutr. Res. 2009, 58, 215–263. [Google Scholar] [CrossRef]
- He, Z.; Zhang, D.; Olanya, O.M. Antioxidant activities of the water-soluble fractions of glandless and glanded cottonseed protein. Food Chem. 2020, 325, 126907. [Google Scholar] [CrossRef]
- He, Z.; Nam, S.; Zhang, H.; Olanya, O.M. Chemical Composition and Thermogravimetric Behaviors of Glanded and Glandless Cottonseed Kernels. Molecules 2022, 27, 316. [Google Scholar] [CrossRef]
- Song, W.; Kong, X.; Hua, Y.; Li, X.; Zhang, C.; Chen, Y. Antioxidant and antibacterial activity and in vitro digestion stability of cottonseed protein hydrolysates. LWT 2020, 118, 108724. [Google Scholar] [CrossRef]
- He, Z.; Zhang, D.; Mattison, C.P. Quantitative comparison of the storage protein distribution in glandless and glanded cottonseeds. Agric. Environ. Lett. 2022, 7, e20076. [Google Scholar] [CrossRef]
- He, Z.; Liu, S.; Nam, S.; Klasson, K.T.; Cheng, H.N. Molecular level characterization of the effect of roasting on the extractable components of glandless cottonseed by Fourier transform ion cyclotron resonance mass spectrometry. Food Chem. 2022, 403, 134404. [Google Scholar] [CrossRef]
- Randel, R.D.; Chase, C.C., Jr.; Wyse, S.J. Effects of gossypol and cottonseed products on reproduction of mammals. J. Anim. Sci. 1992, 70, 1628–1638. [Google Scholar] [CrossRef]
- Zhang, L.M.; Zhang, Y.Z.; Liu, Y.Q.; Gong, Z.H.; Zhao, Y.M.; Li, Y.F. CTN-986, a compound extracted from cottonseeds, increases cell proliferation in hippocampus in vivo and in cultured neural progenitor cells in vitro. Eur. J. Pharmacol. 2009, 607, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Sethumadhavan, K. Cottonseed extracts and gossypol regulate diacylglycerol acyltransferase gene expression in mouse macrophages. J. Agric. Food Chem. 2018, 66, 6022–6030. [Google Scholar] [CrossRef]
- Kong, X.; Song, W.; Hua, Y.; Li, X.; Chen, Y.; Zhang, C.; Chen, Y. Insights into the antibacterial activity of cottonseed protein-derived peptide against Escherichia coli. Food Funct. 2020, 11, 10047–10057. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, M.; Yu, Z.; Du, S.K. Preparation and identification of antioxidant peptides from cottonseed proteins. Food Chem. 2021, 352, 129399. [Google Scholar] [CrossRef]
- Cao, H.; Qin, B.; Panickar, K.S.; Anderson, R.A. Tea and cinnamon polyphenols improve the metabolic syndrome. Agro Food Ind. Hi-Tech 2008, 19, 14–17. [Google Scholar]
- Hu, Q.; Liao, W.; Zhang, Z.; Shi, S.; Hou, S.; Ji, N.; Zhang, X.; Zhang, Q.; Liao, Y.; Li, L.; et al. The hepatoprotective effects of plant-based foods based on the “gut-liver axis”: A prospective review. Crit. Rev. Food Sci. Nutr. 2022, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Hazafa, A.; Rehman, K.U.; Jahan, N.; Jabeen, Z. The Role of Polyphenol (Flavonoids) Compounds in the Treatment of Cancer Cells. Nutr. Cancer 2020, 72, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Guan, P.; Hu, X.; Yang, L.; He, L.; Lin, Q.; Luo, F.; Li, J.; He, X.; Du, Z.; et al. Natural Polyphenols as Targeted Modulators in Colon Cancer: Molecular Mechanisms and Applications. Front. Immunol. 2021, 12, 635484. [Google Scholar] [CrossRef] [PubMed]
- Mileo, A.M.; Nistico, P.; Miccadei, S. Polyphenols: Immunomodulatory and Therapeutic Implication in Colorectal Cancer. Front. Immunol. 2019, 10, 729. [Google Scholar] [CrossRef]
- Kenar, J.A. Reaction chemistry of gossypol and its derivatives. J. Am. Oil Chem. Soc. 2006, 83, 269–302. [Google Scholar] [CrossRef]
- Liu, S.; Kulp, S.K.; Sugimoto, Y.; Jiang, J.; Chang, H.L.; Dowd, M.K.; Wan, P.; Lin, Y.C. The (-)-enantiomer of gossypol possesses higher anticancer potency than racemic gossypol in human breast cancer. Anticancer Res. 2002, 22, 33–38. [Google Scholar] [PubMed]
- Messeha, S.S.; Zarmouh, N.O.; Mendonca, P.; Alwagdani, H.; Cotton, C.; Soliman, K.F.A. Effects of gossypol on apoptosis-related gene expression in racially distinct triple-negative breast cancer cells. Oncol. Rep. 2019, 42, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Leong, J.; Ye, W.; Xu, P.; Lin, S.H.; Liu, J.Y.; Lin, Y.C. (-)-Gossypol-enriched cottonseed oil inhibits proliferation and adipogenesis of human breast pre-adipocytes. Anticancer Res. 2013, 33, 949–955. [Google Scholar] [PubMed]
- Chien, C.C.; Ko, C.H.; Shen, S.C.; Yang, L.Y.; Chen, Y.C. The role of COX-2/PGE2 in gossypol-induced apoptosis of colorectal carcinoma cells. J. Cell Physiol. 2012, 227, 3128–3137. [Google Scholar] [CrossRef]
- Cao, H.; Sethumadhavan, K.; Cao, F.; Wang, T.T.Y. Gossypol decreased cell viability and down-regulated the expression of a number of genes in human colon cancer cells. Sci. Rep. 2021, 11, 5922. [Google Scholar] [CrossRef]
- Yuan, Y.; Tang, A.J.; Castoreno, A.B.; Kuo, S.Y.; Wang, Q.; Kuballa, P.; Xavier, R.; Shamji, A.F.; Schreiber, S.L.; Wagner, B.K. Gossypol and an HMT G9a inhibitor act in synergy to induce cell death in pancreatic cancer cells. Cell Death Dis. 2013, 4, e690. [Google Scholar] [CrossRef] [Green Version]
- Thakur, A.; Lum, L.G.; Schalk, D.; Azmi, A.; Banerjee, S.; Sarkar, F.H.; Mohommad, R. Pan-Bcl-2 inhibitor AT-101 enhances tumor cell killing by EGFR targeted T cells. PLoS ONE 2012, 7, e47520. [Google Scholar] [CrossRef] [Green Version]
- Pang, X.; Wu, Y.; Wu, Y.; Lu, B.; Chen, J.; Wang, J.; Yi, Z.; Qu, W.; Liu, M. (-)-Gossypol suppresses the growth of human prostate cancer xenografts via modulating VEGF signaling-mediated angiogenesis. Mol. Cancer Ther. 2011, 10, 795–805. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.W.; Wang, L.S.; Dowd, M.K.; Wan, P.J.; Lin, Y.C. (-)-Gossypol reduces invasiveness in metastatic prostate cancer cells. Anticancer Res. 2009, 29, 2179–2188. [Google Scholar]
- Huo, M.; Gao, R.; Jiang, L.; Cui, X.; Duan, L.; Deng, X.; Guan, S.; Wei, J.; Soromou, L.W.; Feng, H.; et al. Suppression of LPS-induced inflammatory responses by gossypol in RAW 264.7 cells and mouse models. Int. Immunopharmacol. 2013, 15, 442–449. [Google Scholar] [CrossRef]
- Mellon, J.E.; Zelaya, C.A.; Dowd, M.K.; Beltz, S.B.; Klich, M.A. Inhibitory effects of gossypol, gossypolone, and apogossypolone on a collection of economically important filamentous fungi. J. Agric. Food Chem. 2012, 60, 2740–2745. [Google Scholar] [CrossRef] [PubMed]
- Prior, R.L.; Gu, L. Occurrence and biological significance of proanthocyanidins in the American diet. Phytochemistry 2005, 66, 2264–2280. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Hininger-Favier, I.; Kelly, M.A.; Benaraba, R.; Dawson, H.D.; Coves, S.; Roussel, A.M.; Anderson, R.A. Green tea polyphenol extract regulates the expression of genes involved in glucose uptake and insulin signaling in rats fed a high fructose diet. J. Agric. Food Chem. 2007, 55, 6372–6378. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Kelly, M.A.; Kari, F.; Dawson, H.D.; Urban, J.F., Jr.; Coves, S.; Roussel, A.M.; Anderson, R.A. Green tea increases anti-inflammatory tristetraprolin and decreases pro-inflammatory tumor necrosis factor mRNA levels in rats. J. Inflamm. 2007, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Anderson, R.A. Cinnamon polyphenol extract regulates tristetraprolin and related gene expression in mouse adipocytes. J. Agric. Food Chem. 2011, 59, 2739–2744. [Google Scholar] [CrossRef]
- Cao, H.; Graves, D.J.; Anderson, R.A. Cinnamon extract regulates glucose transporter and insulin-signaling gene expression in mouse adipocytes. Phytomedicine 2010, 17, 1027–1032. [Google Scholar] [CrossRef]
- Cao, H.; Polansky, M.M.; Anderson, R.A. Cinnamon extract and polyphenols affect the expression of tristetraprolin, insulin receptor, and glucose transporter 4 in mouse 3T3-L1 adipocytes. Arch. Biochem. Biophys. 2007, 459, 214–222. [Google Scholar] [CrossRef]
- Cao, H.; Urban, J.F., Jr.; Anderson, R.A. Cinnamon polyphenol extract affects immune responses by regulating anti- and proinflammatory and glucose transporter gene expression in mouse macrophages. J. Nutr. 2008, 138, 833–840. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Sethumadhavan, K.; Li, K.; Boue, S.M.; Anderson, R.A. Cinnamon polyphenol extract and insulin regulate diacylglycerol acyltransferase gene expression in mouse adipocytes and macrophages. Plant Foods Hum. Nutr. 2019, 74, 115–121. [Google Scholar] [CrossRef]
- Cao, H.; Sethumadhavan, K. Gossypol but not cottonseed extracts or lipopolysaccharides stimulates HuR gene expression in mouse cells. J. Funct. Foods 2019, 59, 25–29. [Google Scholar] [CrossRef]
- Cao, H.; Sethumadhavan, K. Regulation of cell viability and anti-inflammatory tristetraprolin family gene expression in mouse macrophages by cottonseed extracts. Sci. Rep. 2020, 10, 775. [Google Scholar] [CrossRef] [Green Version]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Mamat, U.; Schmidt, G.; Loppnow, H.; Ulmer, A.J.; Zähringer, U.; Seydel, U.; Di Padova, F.; et al. Bacterial endotoxin: Molecular relationships of structure to activity and function. FASEB J. 1994, 8, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Cao, F.; Klasson, K.T. Characterization of reference gene expression in tung tree (Vernicia fordii). Ind. Crops Prod. 2013, 50, 248–255. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [Green Version]
- Lejeune, P.; Reisser, D.; Onier, N.; Lagadec, P.; Lindley, I.; Jeannin, J.F. Interleukin-8 has antitumor effects in the rat which are not associated with polymorphonuclear leukocyte cytotoxicity. Cancer Immunol. Immunother. 1994, 38, 167–170. [Google Scholar] [CrossRef]
- Yoshioka, T.; Morimoto, Y.; Iwagaki, H.; Itoh, H.; Saito, S.; Kobayashi, N.; Yagi, T.; Tanaka, N. Bacterial lipopolysaccharide induces transforming growth factor beta and hepatocyte growth factor through toll-like receptor 2 in cultured human colon cancer cells. J. Int. Med. Res. 2001, 29, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Ikebe, M.; Kitaura, Y.; Nakamura, M.; Tanaka, H.; Yamasaki, A.; Nagai, S.; Wada, J.; Yanai, K.; Koga, K.; Sato, N.; et al. Lipopolysaccharide (LPS) increases the invasive ability of pancreatic cancer cells through the TLR4/MyD88 signaling pathway. J. Surg.Oncol. 2009, 100, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.T.; Jing, Y.Y.; Yan, F.; Han, Z.P.; Lai, F.B.; Zeng, J.X.; Yu, G.F.; Fan, Q.M.; Li, R.; Zhao, Q.D.; et al. LPS-induced CXCR4-dependent migratory properties and a mesenchymal-like phenotype of colorectal cancer cells. Cell Adhes. Migr. 2017, 11, 13–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.H.; Kim, W.T.; Kim, D.H.; Park, J.W.; Kang, T.H.; Chung, J.W.; Leem, S.H. Tristetraprolin suppresses AHRR expression through mRNA destabilization. FEBS Lett. 2013, 587, 1518–1523. [Google Scholar] [CrossRef] [Green Version]
- Kitada, S.; Kress, C.L.; Krajewska, M.; Jia, L.; Pellecchia, M.; Reed, J.C. Bcl-2 antagonist apogossypol (NSC736630) displays single-agent activity in Bcl-2-transgenic mice and has superior efficacy with less toxicity compared with gossypol (NSC19048). Blood 2008, 111, 3211–3219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frevel, M.A.; Bakheet, T.; Silva, A.M.; Hissong, J.G.; Khabar, K.S.; Williams, B.R. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 2003, 23, 425–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Ning, H.; Gu, L.; Wang, Q.; Lu, W.; Peng, H.; Cui, W.; Ying, B.; Ross, C.R.; Wilson, G.M.; et al. Tristetraprolin induces cell cycle arrest in breast tumor cells through targeting AP-1/c-Jun and NF-kappaB pathway. Oncotarget 2015, 6, 41679–41691. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Bhat, A.A.; Krishnan, M.; Singh, A.B.; Dhawan, P. Trichostatin-A modulates claudin-1 mRNA stability through the modulation of Hu antigen R and tristetraprolin in colon cancer cells. Carcinogenesis 2013, 34, 2610–2621. [Google Scholar] [CrossRef] [Green Version]
- Warzych, E.; Wolc, A.; Cieslak, A.; Lechniak-Cieslak, D. 217 transcript abundance of cathepsin genes in cumulus cells as a marker of cattle oocyte quality. Reprod. Fertil. Dev. 2012, 25, 257. [Google Scholar] [CrossRef]
- Sawaoka, H.; Dixon, D.A.; Oates, J.A.; Boutaud, O. Tristetraprolin binds to the 3’-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J. Biol. Chem. 2003, 278, 13928–13935. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.H.; Lee, H.H.; Vo, M.T.; Kim, H.J.; Ko, M.S.; Im, Y.C.; Min, Y.J.; Lee, B.J.; Cho, W.J.; Park, J.W. Casein kinase 2 regulates the mRNA-destabilizing activity of tristetraprolin. J. Biol. Chem. 2011, 286, 21577–21587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrmann, D.C.; Tausendschon, M.; Wittig, I.; Steger, M.; Ding, M.G.; Schmid, T.; Dehne, N.; Brune, B. Inactivation of tristetraprolin in chronic hypoxia provokes the expression of cathepsin B. Mol. Cell Biol. 2015, 35, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Biswas, R.; Novotny, M.; Pavicic, P.G., Jr.; Herjan, T.; Mandal, P.; Hamilton, T.A. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J. Immunol. 2008, 180, 2545–2552. [Google Scholar] [CrossRef] [Green Version]
- Ligueros, M.; Jeoung, D.; Tang, B.; Hochhauser, D.; Reidenberg, M.M.; Sonenberg, M. Gossypol inhibition of mitosis, cyclin D1 and Rb protein in human mammary cancer cells and cyclin-D1 transfected human fibrosarcoma cells. Br. J. Cancer 1997, 76, 21–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Mao, B.; Li, L.; Guan, H.; Su, Y.; Li, X.; Lian, Q.; Huang, P.; Ge, R.S. Gossypol enantiomers potently inhibit human placental 3beta-hydroxysteroid dehydrogenase 1 and aromatase activities. Fitoterapia 2015, 109, 132–137. [Google Scholar] [CrossRef]
- Ludwig, E.H.; Mahley, R.W.; Palaoglu, E.; Ozbayrakci, S.; Balestra, M.E.; Borecki, I.B.; Innerarity, T.L.; Farese, R.V., Jr. DGAT1 promoter polymorphism associated with alterations in body mass index, high density lipoprotein levels and blood pressure in Turkish women. Clin. Genet. 2002, 62, 68–73. [Google Scholar] [CrossRef]
- Dey, P.; Chakraborty, M.; Kamdar, M.R.; Maiti, M.K. Functional characterization of two structurally novel diacylglycerol acyltransferase2 isozymes responsible for the enhanced production of stearate-rich storage lipid in Candida tropicalis SY005. PLoS ONE 2014, 9, e94472. [Google Scholar] [CrossRef]
- Lee, H.H.; Lee, S.R.; Leem, S.H. Tristetraprolin regulates prostate cancer cell growth through suppression of E2F1. J. Microbiol. Biotechnol. 2014, 24, 287–294. [Google Scholar] [CrossRef] [Green Version]
- Florkowska, M.; Tymoszuk, P.; Balwierz, A.; Skucha, A.; Kochan, J.; Wawro, M.; Stalinska, K.; Kasza, A. EGF activates TTP expression by activation of ELK-1 and EGR-1 transcription factors. BMC Mol. Biol. 2012, 13, 8. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.S.; Hsu, Y.L.; Kuo, P.L.; Chiang, L.C.; Lin, C.C. Upregulation of Fas/Fas ligand-mediated apoptosis by gossypol in an immortalized human alveolar lung cancer cell line. Clin. Exp. Pharmacol. Physiol. 2004, 31, 716–722. [Google Scholar] [CrossRef]
- Chen, D.; Pan, X.; Xiao, P.; Farwell, M.A.; Zhang, B. Evaluation and identification of reliable reference genes for pharmacogenomics, toxicogenomics, and small RNA expression analysis. J. Cell Physiol. 2011, 226, 2469–2477. [Google Scholar] [CrossRef]
- Fahling, M.; Persson, A.B.; Klinger, B.; Benko, E.; Steege, A.; Kasim, M.; Patzak, A.; Persson, P.B.; Wolf, G.; Bluthgen, N.; et al. Multilevel regulation of HIF-1 signaling by TTP. Mol. Biol. Cell 2012, 23, 4129–4141. [Google Scholar] [CrossRef] [PubMed]
- Kagami, S.; Kanari, H.; Suto, A.; Fujiwara, M.; Ikeda, K.; Hirose, K.; Watanabe, N.; Iwamoto, I.; Nakajima, H. HMG-CoA reductase inhibitor simvastatin inhibits proinflammatory cytokine production from murine mast cells. Int. Arch. Allergy Immunol. 2008, 146, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Jamal, U.M.; Joe, Y.; Zheng, M.; Blackshear, P.J.; Ryter, S.W.; Park, J.W.; Chung, H.T. A functional link between heme oxygenase-1 and tristetraprolin in the anti-inflammatory effects of nicotine. Free Radic. Biol. Med. 2013, 65, 1331–1339. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.X.; Li, J.S.; Hu, R.; Shi, Y.; Su, X.; Li, Q.; Zhang, F. CNOT7/hCAF1 is involved in ICAM-1 and IL-8 regulation by tristetraprolin. Cell Signal. 2014, 26, 2390–2396. [Google Scholar] [CrossRef] [PubMed]
- Su, N.Y.; Tsai, P.S.; Huang, C.J. Clonidine-Induced Enhancement of iNOS Expression Involves NF-kappaB. J. Surg. Res. 2008, 149, 131–137. [Google Scholar] [CrossRef]
- Ogilvie, R.L.; Abelson, M.; Hau, H.H.; Vlasova, I.; Blackshear, P.J.; Bohjanen, P.R. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005, 174, 953–961. [Google Scholar] [CrossRef] [Green Version]
- Hochdorfer, T.; Tiedje, C.; Stumpo, D.J.; Blackshear, P.J.; Gaestel, M.; Huber, M. LPS-induced production of TNF-alpha and IL-6 in mast cells is dependent on p38 but independent of TTP. Cell Signal. 2013, 25, 1339–1347. [Google Scholar] [CrossRef] [Green Version]
- Balakathiresan, N.S.; Bhattacharyya, S.; Gutti, U.; Long, R.P.; Jozwik, C.; Huang, W.; Srivastava, M.; Pollard, H.B.; Biswas, R. Tristetraprolin regulates IL-8 mRNA stability in cystic fibrosis lung epithelial cells. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2009, 296, L1012–L1018. [Google Scholar] [CrossRef] [Green Version]
- Gaba, A.; Grivennikov, S.I.; Do, M.V.; Stumpo, D.J.; Blackshear, P.J.; Karin, M. Cutting edge: IL-10-mediated tristetraprolin induction is part of a feedback loop that controls macrophage STAT3 activation and cytokine production. J. Immunol. 2012, 189, 2089–2093. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.; Ning, H.; Qian, X.; Huang, Q.; Hou, R.; Almourani, R.; Fu, M.; Blackshear, P.J.; Liu, J. Suppression of IL-12 production by tristetraprolin through blocking NF-kcyB nuclear translocation. J. Immunol. 2013, 191, 3922–3930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milke, L.; Schulz, K.; Weigert, A.; Sha, W.; Schmid, T.; Brune, B. Depletion of tristetraprolin in breast cancer cells increases interleukin-16 expression and promotes tumor infiltration with monocytes/macrophages. Carcinogenesis 2013, 34, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Novotny, M.; Pavicic, P.G., Jr.; Zhao, C.; Herjan, T.; Hartupee, J.; Hamilton, T. IL-17 regulates CXCL1 mRNA stability via an AUUUA/tristetraprolin-independent sequence. J. Immunol. 2010, 184, 1484–1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Ye, W.; Zhong, S.; Li, H.; Feng, E.; Lin, S.H.; Kuo, C.T.; Liu, J.Y.; Lin, Y.C. Leptin and zeranol up-regulate cyclin D1 expression in primary cultured normal human breast pre-adipocytes. Mol. Med. Rep. 2010, 3, 983–990. [Google Scholar] [CrossRef]
- Voss, A.K.; Thomas, T.; Gruss, P. Compensation for a gene trap mutation in the murine microtubule-associated protein 4 locus by alternative polyadenylation and alternative splicing. Dev. Dyn. 1998, 212, 258–266. [Google Scholar] [CrossRef]
- Jiang, J.; Slivova, V.; Jedinak, A.; Sliva, D. Gossypol inhibits growth, invasiveness, and angiogenesis in human prostate cancer cells by modulating NF-kappaB/AP-1 dependent- and independent-signaling. Clin. Exp. Metastasis 2012, 29, 165–178. [Google Scholar] [CrossRef]
- Barba-Barajas, M.; Hernandez-Flores, G.; Lerma-Diaz, J.M.; Ortiz-Lazareno, P.C.; Dominguez-Rodriguez, J.R.; Barba-Barajas, L.; de Celis, R.; Jave-Suarez, L.F.; Aguilar-Lemarroy, A.C.; Guevara-Barraza, M.G.; et al. Gossypol induced apoptosis of polymorphonuclear leukocytes and monocytes: Involvement of mitochondrial pathway and reactive oxygen species. Immunopharmacol. Immunotoxicol. 2009, 31, 320–330. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, C.W.; Vo, M.T.; Lee, H.H.; Lee, J.Y.; Yoon, N.A.; Lee, C.Y.; Moon, C.H.; Min, Y.J.; Park, J.W.; et al. Expression of proviral integration site for Moloney murine leukemia virus 1 (Pim-1) is post-transcriptionally regulated by tristetraprolin in cancer cells. J. Biol. Chem. 2012, 287, 28770–28778. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.W.; Wang, L.S.; Chang, H.L.; Ye, W.; Dowd, M.K.; Wan, P.J.; Lin, Y.C. Molecular mechanisms of (-)-gossypol-induced apoptosis in human prostate cancer cells. Anticancer Res. 2006, 26, 1925–1933. [Google Scholar]
- Militello, R.D.; Munafo, D.B.; Beron, W.; Lopez, L.A.; Onier, S.; Oud, B.; Olombo, M.I.; Oloris, S.C. Rab24 is Required for Normal Cell Division. Traffic 2013, 14, 502–518. [Google Scholar] [CrossRef]
- Brattelid, T.; Winer, L.H.; Levy, F.O.; Liestol, K.; Sejersted, O.M.; Andersson, K.B. Reference gene alternatives to Gapdh in rodent and human heart failure gene expression studies. BMC Mol. Biol. 2010, 11, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeow, W.S.; Baras, A.; Chua, A.; Nguyen, D.M.; Sehgal, S.S.; Schrump, D.S.; Nguyen, D.M. Gossypol, a phytochemical with BH3-mimetic property, sensitizes cultured thoracic cancer cells to Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J. Thorac. Cardiovasc. Surg. 2006, 132, 1356–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Shen, Z.; Shang, L.; Wang, X. Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death. Differ. 2011, 18, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Essafi-Benkhadir, K.; Onesto, C.; Stebe, E.; Moroni, C.; Pages, G. Tristetraprolin inhibits ras-dependent tumor vascularization by inducing VEGF mRNA degradation. Mol. Biol. Cell 2007, 18, 4648–4658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, G.; Sun, D.; Ou, Z.; Ding, A. The protein Zfand5 binds and stabilizes mRNAs with AU-rich elements in their 3’-untranslated regions. J. Biol. Chem. 2012, 287, 24967–24977. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Shockey, J.M. Comparison of TaqMan and SYBR Green qPCR methods for quantitative gene expression in tung tree tissues. J. Agric. Food Chem. 2012, 60, 12296–12303. [Google Scholar] [CrossRef]
- Yun, E.J.; Yu, S.; Kim, Y.A.; Liu, J.J.; Kang, N.J.; Jin, Y.S.; Kim, K.H. In Vitro Prebiotic and Anti-Colon Cancer Activities of Agar-Derived Sugars from Red Seaweeds. Mar. Drugs 2021, 19, 213. [Google Scholar] [CrossRef]
- Zhao, X.; Song, J.L.; Kim, J.D.; Lee, J.S.; Park, K.Y. Fermented Pu-erh tea increases in vitro anticancer activities in HT-29 cells and has antiangiogenetic effects on HUVECs. J. Environ. Pathol. Toxicol. Oncol. 2013, 32, 275–288. [Google Scholar] [CrossRef]
- Gu, Q.; Hu, C.; Chen, Q.; Xia, Y. Tea polyphenols prevent lung from preneoplastic lesions and effect p53 and bcl-2 gene expression in rat lung tissues. Int. J. Clin. Exp. Pathol. 2013, 6, 1523–1531. [Google Scholar]
- Sandur, S.K.; Deorukhkar, A.; Pandey, M.K.; Pabon, A.M.; Shentu, S.; Guha, S.; Aggarwal, B.B.; Krishnan, S. Curcumin modulates the radiosensitivity of colorectal cancer cells by suppressing constitutive and inducible NF-kappaB activity. Int. J. Radiat. Oncol. Biol. Phys. 2009, 75, 534–542. [Google Scholar] [CrossRef] [Green Version]
- Su, C.C.; Lin, J.G.; Li, T.M.; Chung, J.G.; Yang, J.S.; Ip, S.W.; Lin, W.C.; Chen, G.W. Curcumin-induced apoptosis of human colon cancer colo 205 cells through the production of ROS, Ca2+ and the activation of caspase-3. Anticancer Res. 2006, 26, 4379–4389. [Google Scholar] [PubMed]
- Laka, K.; Mapheto, K.B.F.; Mbita, Z. Selective in vitro cytotoxicity effect of Drimia calcarata bulb extracts against p53 mutant HT-29 and p53 wild-type Caco-2 colorectal cancer cells through STAT5B regulation. Toxicol. Rep. 2021, 8, 1265–1279. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Mu, L.; Cui, Y.; Li, Y.; Chen, P.; Xie, H.; Wang, X. Berberine Promotes Apoptosis of Colorectal Cancer via Regulation of the Long Non-Coding RNA (lncRNA) Cancer Susceptibility Candidate 2 (CASC2)/AU-Binding Factor 1 (AUF1)/B-Cell CLL/Lymphoma 2 (Bcl-2) Axis. Med. Sci. Monit. 2019, 25, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.H.; Shen, S.C.; Yang, L.Y.; Lin, C.W.; Chen, Y.C. Gossypol reduction of tumor growth through ROS-dependent mitochondria pathway in human colorectal carcinoma cells. Int. J. Cancer 2007, 121, 1670–1679. [Google Scholar] [CrossRef]
- Lan, L.; Appelman, C.; Smith, A.R.; Yu, J.; Larsen, S.; Marquez, R.T.; Liu, H.; Wu, X.; Gao, P.; Roy, A.; et al. Natural product (-)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol.Oncol. 2015, 9, 1406–1420. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Wang, J.; Cheng, G.; Zhu, X.F.; Huang, P.; Yang, D.; Zeng, Y.X. Apogossypolone targets mitochondria and light enhances its anticancer activity by stimulating generation of singlet oxygen and reactive oxygen species. Chin. J. Cancer 2011, 30, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Cao, X.X.; Jiang, H.X.; Zhao, X.L.; Wang, J.Y.; Lin, Y.H.; Liu, Q.L.; Zhang, C.; Jiang, B.; Guo, F. A novel water-soluble gossypol derivative increases chemotherapeutic sensitivity and promotes growth inhibition in colon cancer. J. Med. Chem. 2010, 53, 5502–5510. [Google Scholar] [CrossRef]
- Cao, H.; Cao, F.; Roussel, A.M.; Anderson, R.A. Quantitative PCR for glucose transporter and tristetraprolin family gene expression in cultured mouse adipocytes and macrophages. In Vitro Cell Dev. Biol. Anim. 2013, 49, 759–770. [Google Scholar] [CrossRef]








| ID | mRNA | Name | Forward Primer (5' to 3') | Reverse Primer (5' to 3') | Regulator [Reference] |
|---|---|---|---|---|---|
| H1 | Ahrr1 | Aryl hydrocarbon receptor repressor | AGGCTGCTGTTGGAGTCTCTTAA | CGATCGTTGCTGATGCATAAA | TTP [55] |
| H2 | Bcl2 | B-cell lymphoma 2 | CAGCATGCGGCCTCTGTT | GGGCCAAACTGAGCAGAGTCT | Gossypol [56] |
| H3 | Bcl2l1 | B-cell lymphoma 2 like 1 | GTGCGTGGAAAGCGTAGACA | ATTCAGGTAAGTGGCCATCCAA | TTP [57] |
| H4 | Bnip3 | BCL2 protein-interacting protein 3 | GTCAAGTCGGCCGGAAAATA | TGCGCTTCGGGTGTTTAAAG | Gossypol [27] |
| H5 | Cd36 | Cluster of differentiation 36/fatty acid translocase | CTCTTTCCTGCAGCCCAATG | TTGTCAGCCTCTGTTCCAACTG | TTP [58] |
| H6 | Claudin1 | Maintain tissue integrity and water retention | GACAAAGTGAAGAAGGCCCGTAT | CAAGACCTGCCACGATGAAA | TTP [59] |
| H7 | Cox1 | Cyclooxygenase 1 | CGCCCACGCCAGTGA | AGGCCGAAGCGGACACA | TTP [60] |
| H8 | Cox2 | Cyclooxygenase 2 | CGATTGTACCCGGACAGGAT | TTGGAGTGGGTTTCAGAAATAATTT | TTP [61] |
| H9 | Csnk2a1 | Casein kinase 2 alpha 1 | AGCGATGGGAACGCTTTG | AAGGCCTCAGGGCTGACAA | TTP [62] |
| H10 | Ctsb | Cathepsin B | GACTTGTAGCTGCTGTCTCTCTTTGT | CAAGAGTCGCAAGAACATGCA | TTP [63] |
| H11 | Cxcl1 | Chemokine (C-X-C motif) ligand 1 | GCCCAAACCGAAGTCATAGC | TGCAGGATTGAGGCAAGCT | TTP [64] |
| H12 | Cyclind1 | Cyclin D1 | ACACGCGCAGACCTTCGT | CCATGGAGGGCGGATTG | Gossypol [65] |
| H13 | Cyp19a1 | Cytochrome P450 family 19 subfamily A member 1 | GACATTGCAAGGACAGTGTGTTG | AGTCTCATCTGGGTGCAAGGA | Gossypol [66] |
| H14 | Dgat1 | Diacylglycerol O-acyltransferase 1 | ACCTCATCTGGCTCATCTTCTTCTA | CCCGGTCTCCAAACTGCAT | Cinnamon [40,67] |
| H15 | Dgat2a | Diacylglycerol O-acyltransferase 2a | CCCAGGCATACGGCCTTA | CAACACAGGCATTCGGAAGTT | Cinnamon [40,68] |
| H16 | Dgat2b | Diacylglycerol O-acyltransferase 2b | ACTCTGGCCCTTCTCTGTTTTTTA | TCCACCTTGGTTGGGTGTGT | Cinnamon [40,68] |
| H17 | E2f1 | E2F transcription factor 1 | CGGCGCATCTATGACATCAC | CAGCCACTGGATGTGGTTCTT | TTP [69] |
| H18 | Elk1 | ETS transcription factor | CTCCTCCGCATCCCTCTTTAA | AGCGTCACAGATGGGTCCAT | TTP [70] |
| H19 | Fas | Fas cell surface death receptor | GAACTCCTTGGCGGAAGAGA | AGGACCCCGTGGAATGTCA | Gossypol [71] |
| H20 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | GGGTGTGAACCATGAGAAGTATGA | GGTGCAGGAGGCATTGCT | [72] |
| H21 | Glut1 | Glucose transporter 1 | TGCTCATGGGCTTCTCGAA | AAGCGGCCCAGGATCAG | Cinnamon [39] |
| H22 | Glut2 | Glucose transporter 2 | GCATTTTTCAGACGGCTGGTA | GCGCCAACTCCAATGGTT | Cinnamon [39] |
| H23 | Glut3 | Glucose transporter 3 | GAGGATATCACACGGGCCTTT | CCATGACGCCGTCCTTTC | Cinnamon [39] |
| H24 | Glut4 | Glucose transporter 4 | CGTGGGCGGCATGATT | CCAGCATGGCCCTTTTCC | Cinnamon [39] |
| H25 | Hif1a | Hypoxia inducible factor 1 subunit alpha | GGTGGATATGTCTGGGTTGAAAC | ATGCACTGTGGTTGAGAATTCTTG | TTP [73] |
| H26 | Hmgr | 3-Hydroxy-3-methylglutaryl-CoA reductase | AAGTGAAAGCCTGGCTCGAA | CTAGTGCTGTCAAATGCCTCCTT | [74] |
| H27 | Hmox1 | Heme oxygenase 1 | CTTCTCCGATGGGTCCTTACACT | TCACATGGCATAAAGCCCTACA | TTP [75] |
| H28 | Hua | Human antigen a | GATCCTCTGGCAGATGTTTGG | CGCGGATCACTTTCACATTG | Gossypol [41] |
| H29 | Icam1 | Intercellular adhesion molecule 1/CD54 | GGAGCTTCGTGTCCTGTATGG | TTTCTGGCCACGTCCAGTTT | [76] |
| H30 | Inos | Inducible nitric oxide synthase | AGATCCGGTTCACAGTCTTGGT | GCCATGACCTTCCGCATTAG | [77] |
| H31 | Insr | Insulin receptor | CAACGGGCAGTTTGTCGAA | TGGTCGGGCAAACTTTCTG | [38] |
| H32 | Il2 | Interleukin 2 | TATGCAGATGAGACAGCAACCAT | TTGAGATGATGCTTTGACAAAAGG | TTP [78] |
| H33 | IL6 | Interleukin 6 | CCCACACAGACAGCCACTCA | CCGTCGAGGATGTACCGAAT | TTP [79] |
| H34 | IL8 | Interleukin 8 | CCATCTCACTGTGTGTAAACATGACTT | ATCAGGAAGGCTGCCAAGAG | TTP [80] |
| H35 | Il10 | Interleukin 10 | GCCGTGGAGCAGGTGAAG | TGGCTTTGTAGATGCCTTTCTCT | TTP [81] |
| H36 | Il12 | Interleukin 12 | TGCCTTCACCACTCCCAAA | TGTCTGGCCTTCTGGAGCAT | TTP [82] |
| H37 | Il16 | Interleukin 16 | CAGGGCCTCACACGGTTT | GACAATCGTGACAGGTCCATCA | TTP [83] |
| H38 | Il17 | Interleukin 17 | CCCAAAAGGTCCTCAGATTACTACA | TCATTGCGGTGGAGATTCC | TTP [84] |
| H39 | Leptin | Body fat and obesity hormone | AGGGAGACCGAGCGCTTT | CACATCCCTCACCTCCTTCAAA | [85] |
| H40 | Map1lc3a | Microtubule-associated proteins 1 light chain 3A | GTGAACCAGCACAGCATGGT | CCTCGTCTTTCTCCTGCTCGTA | [86] |
| H41 | Map1lc3b | Microtubule-associated proteins 1 light chain 3B | AGGCGCTTACAGCTCAATGC | ACCATGCTGTGTCCGTTCAC | [86] |
| H42 | Nfkb | Nuclear factor kappa B | GGTGCCTCTAGTGAAAAGAACAAGA | GCTGGTCCCACATAGTTGCA | [87] |
| H43 | P53 | Tumor suppressor | CTTGCAATAGGTGTGCGTCAGA | GGAGCCCCGGGACAAA | Gossypol [88] |
| H44 | Pim1 | Proto-oncogene serine/threonine-protein kinase | TGCTCCACCGCGACATC | TGAGCTCGCCGCGATT | TTP [89] |
| H45 | Pparr | Peroxisome proliferator-activated receptor γ | GAACGACCAAGTAACTCTCCTCAAA | CAAGGAGGCCAGCATTGTGT | Gossypol [90] |
| H46 | Rab24 | Ras-related oncogene 24 | TCGGTCGGAGACGCACTT | TGGCCTCATAGCGCTCAGA | [91] |
| H47 | Rpl32 | Ribosomal protein L32 (60S ribosomal unit) | CCTCCAAGAACCGCAAAGC | GGTGACTCTGATGGCCAGTTG | [92] |
| H48 | Tnf | Tumor necrosis factor | GGAGAAGGGTGACCGACTCA | CAGACTCGGCAAAGTCGAGAT | TTP [79] |
| H49 | Tnfsf10 | Tumor necrosis factor superfamily, member 10 | GCTCTGGGCCGCAAAAT | AGGAATGAATGCCCACTCCTT | Gossypol [93] |
| H50 | Ulk2 | Unc-51 like autophagy activating kinase 2 | ACAGCTCCTTTCAAAATCCCTAAA | AGGCCCATGACGAGTAACCA | [94] |
| H51 | Vegf | Vascular endothelial growth factor | CCCACTGAGGAGTCCAACATC | GGCCTTGGTGAGGTTTGATC | TTP [95] |
| H52 | Zfand5 | Zinc finger AN1-type containing 5 | AGGGTTTGACTGCCGATGTG | ACTGGATTCTCTTTTCTGATTTTTGC | TTP [96] |
| H53 | Zfp36/Ttp | Zinc finger protein 36/Tristetraprolin | GGCGACTCCCCATCTTCAA | GACCGGGCAGTCACTTTGTC | TTP [38] |
| H54 | Zfp36l1 | Zinc finger protein 36 like 1 | TCTGCCACCATCTTCGACTTG | TGGGAGCACTATAGTTGAGCATCT | TTP [38] |
| H55 | Zfp36l2 | Zinc finger protein 36 like 2 | CCTTTCATACCATCGGCTTCTG | TCGTCCGCGTTGTGGAT | TTP [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, H.; Sethumadhavan, K. Identification of Bcl2 as a Stably Expressed qPCR Reference Gene for Human Colon Cancer Cells Treated with Cottonseed-Derived Gossypol and Bioactive Extracts and Bacteria-Derived Lipopolysaccharides. Molecules 2022, 27, 7560. https://doi.org/10.3390/molecules27217560
Cao H, Sethumadhavan K. Identification of Bcl2 as a Stably Expressed qPCR Reference Gene for Human Colon Cancer Cells Treated with Cottonseed-Derived Gossypol and Bioactive Extracts and Bacteria-Derived Lipopolysaccharides. Molecules. 2022; 27(21):7560. https://doi.org/10.3390/molecules27217560
Chicago/Turabian StyleCao, Heping, and Kandan Sethumadhavan. 2022. "Identification of Bcl2 as a Stably Expressed qPCR Reference Gene for Human Colon Cancer Cells Treated with Cottonseed-Derived Gossypol and Bioactive Extracts and Bacteria-Derived Lipopolysaccharides" Molecules 27, no. 21: 7560. https://doi.org/10.3390/molecules27217560