Design of Novel IRAK4 Inhibitors Using Molecular Docking, Dynamics Simulation and 3D-QSAR Studies
Abstract
:1. Introduction
2. Results and Discussion
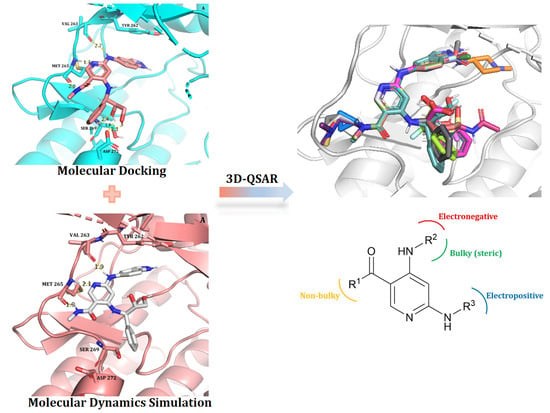
2.1. Molecular Docking
2.2. Molecular Dynamics Simulation
2.3. MM/PBSA Binding Free Energy Calculation
2.4. 3D-QSAR (CoMFA and RF-CoMFA)
Validation of RF-CoMFA Model
2.5. Contour Map Analysis
RF-CoMFA Contour Maps
2.6. Designing of IRAK4 Inhibitors and Their ADMET Calculation
3. Materials and Methods
3.1. Test Set/Training Set Selection for 3D-QSAR Analyses
3.2. Modeling of the Missing Residues
3.3. Preparation of the Protein and Molecular Docking
3.4. Molecular Dynamics Simulations
3.5. MM/PBSA Binding Free Energy Calculations
3.6. CoMFA and RF-CoMFA
Model Validation
3.7. Design of New IRAK4 Inhibitors and ADMET Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kargbo, R.B. PROTAC Degradation of IRAK4 for the Treatment of Cancer. ACS Med. Chem. Lett. 2019, 10, 1370–1371. [Google Scholar] [CrossRef]
- Bhide, R.S.; Keon, A.; Weigelt, C.; Sack, J.S.; Schmidt, R.J.; Lin, S.; Xiao, H.-Y.; Spergel, S.H.; Kempson, J.; Pitts, W.J.; et al. Discovery and structure-based design of 4,6-diaminonicotinamides as potent and selective IRAK4 inhibitors. Bioorgan. Med. Chem. Lett. 2017, 27, 4908–4913. [Google Scholar] [CrossRef]
- Chaudhary, D.; Robinson, S.; Romero, D.L. Recent advances in the discovery of small molecule inhibitors of interleukin-1 receptor-associated kinase 4 (IRAK4) as a therapeutic target for inflammation and oncology disorders: Miniperspective. J. Med. Chem. 2015, 58, 96–110. [Google Scholar] [CrossRef]
- Rhyasen, G.W.; Starczynowski, D.T. IRAK signalling in cancer. Br. J. Cancer 2015, 112, 232–237. [Google Scholar] [CrossRef]
- Warner, N.; Núñez, G. MyD88: A Critical Adaptor Protein in Innate Immunity Signal Transduction. J. Immunol. 2013, 190, 3–4. [Google Scholar] [CrossRef]
- Buckley, G.M.; Gowers, L.; Higueruelo, A.P.; Jenkins, K.; Mack, S.R.; Morgan, T.; Parry, D.M.; Pitt, W.R.; Rausch, O.; Richard, M.D.; et al. IRAK-4 inhibitors. Part 1: A series of amides. Bioorgan. Med. Chem. Lett. 2008, 18, 3211–3214. [Google Scholar] [CrossRef]
- Genung, N.; Guckian, K. Small Molecule Inhibition of Interleukin-1 Receptor-Associated Kinase 4 (IRAK4). Prog. Med. Chem. 2017, 56, 117–163. [Google Scholar] [CrossRef]
- Dunne, A.; Carpenter, S.; Brikos, C.; Gray, P.; Strelow, A.; Wesche, H.; Morrice, N.; O’Neill, L. IRAK1 and IRAK4 Promote Phosphorylation, Ubiquitination, and Degradation of MyD88 Adaptor-like (Mal). J. Biol. Chem. 2010, 285, 18276–18282. [Google Scholar] [CrossRef]
- Flannery, S.; Bowie, A.G. The interleukin-1 receptor-associated kinases: Critical regulators of innate immune signalling. Biochem. Pharmacol. 2010, 80, 1981–1991. [Google Scholar] [CrossRef]
- Medvedev, A.E.; Lentschat, A.; Kuhns, D.B.; Blanco, J.; Salkowski, C.; Zhang, S.; Arditi, M.; Gallin, J.I.; Vogel, S.N. Distinct Mutations in IRAK-4 Confer Hyporesponsiveness to Lipopolysaccharide and Interleukin-1 in a Patient with Recurrent Bacterial Infections. J. Exp. Med. 2003, 198, 521–531. [Google Scholar] [CrossRef]
- Khanfar, M.A.; Alqtaishat, S. Discovery of potent IRAK-4 inhibitors as potential anti-inflammatory and anticancer agents using structure-based exploration of IRAK-4 pharmacophoric space coupled with QSAR analyses. Comput. Biol. Chem. 2019, 79, 147–154. [Google Scholar] [CrossRef]
- Srivastava, R.; Geng, D.; Liu, Y.; Zheng, L.; Li, Z.; Joseph, M.A.; McKenna, C.; Bansal, N.; Ochoa, A.; Davila, E. Augmentation of Therapeutic Responses in Melanoma by Inhibition of IRAK-1,-4IRAK-4 Signaling and Melanoma Progression. Cancer Res. 2012, 72, 6209–6216. [Google Scholar] [CrossRef]
- Khurana, N.; Dodhiawala, P.B.; Bulle, A.; Lim, K.-H. Deciphering the role of innate immune NF-ĸB pathway in pancreatic cancer. Cancers 2020, 12, 2675. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Li, Q.; Chen, Y.; Zhang, D.; Grossman, J.; Li, L.; Khurana, N.; Jiang, H.; Grierson, P.M.; Herndon, J.; DeNardo, D.G.; et al. IRAK4 mediates colitis-induced tumorigenesis and chemoresistance in colorectal cancer. JCI Insight 2019, 4, 130867. [Google Scholar] [CrossRef]
- Poso, A.; von Wright, A.; Gynther, J. An empirical and theoretical study on mechanisms of mutagenic activity of hydrazine compounds. Mutat. Res. Mol. Mech. Mutagen. 1995, 332, 63–71. [Google Scholar] [CrossRef]
- Seganish, W.M. Inhibitors of interleukin-1 receptor-associated kinase 4 (IRAK4): A patent review (2012–2015). Expert Opin. Ther. Patents 2016, 26, 917–932. [Google Scholar] [CrossRef]
- McElroy, W.T. Interleukin-1 receptor-associated kinase 4 (IRAK4) inhibitors: An updated patent review (2016–2018). Expert Opin. Ther. Pat. 2019, 29, 243–259. [Google Scholar] [CrossRef]
- Eisenberg, D.; Schwarz, E.; Komaromy, M.; Wall, R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984, 179, 125–142. [Google Scholar] [CrossRef]
- Lemkul, J. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package [Article v1.0]. Living J. Comput. Mol. Sci. 2018, 1, 5068. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Consortium, O.S.D.D.; Lynn, A.J. modeling. g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Modeling 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. Mol. Divers. 2000, 5, 231–243. [Google Scholar] [CrossRef]
- Gadhe, C.G.; Kothandan, G.; Cho, S.J. Large variation in electrostatic contours upon addition of steric parameters and the effect of charge calculation schemes in CoMFA on mutagenicity of MX analogues. Mol. Simul. 2012, 38, 861–871. [Google Scholar] [CrossRef]
- Roy, K.; Chakraborty, P.; Mitra, I.; Ojha, P.K.; Kar, S.; Das, R.N. Some case studies on application of “rm2” metrics for judging quality of quantitative structure–activity relationship predictions: Emphasis on scaling of response data. J. Comput. Chem. 2013, 34, 1071–1082. [Google Scholar] [CrossRef]
- Thibaut, U.; Folkers, G.; Klebe, G.; Kubinyi, H.; Merz, A.; Rognan, D. Recommendations for CoMFA Studies and 3D QSAR Publications. Quant. Struct. Relatsh. 1994, 13, 1–3. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein structure modeling with MODELLER. In Structural Genomics; Springer: Berlin/Heidelberg, Germany, 2021; pp. 239–255. [Google Scholar]
- Melo, F.; Sánchez, R.; Sali, A. Statistical potentials for fold assessment. Protein Sci. 2002, 11, 430–448. [Google Scholar] [CrossRef]
- Shen, M.-Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.; Olson, A.J.; Goodsell, D.S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Sousa da Silva, A.W.; Vranken, W.F. ACPYPE-Antechamber python parser interface. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Gohlke, H.; Kiel, C.; Case, D.A. Insights into Protein–Protein Binding by Binding Free Energy Calculation and Free Energy Decomposition for the Ras–Raf and Ras–RalGDS Complexes. J. Mol. Biol. 2003, 330, 891–913. [Google Scholar] [CrossRef]
- Chirico, N.; Gramatica, P. Real External Predictivity of QSAR Models. Part 2. New Intercomparable Thresholds for Different Validation Criteria and the Need for Scatter Plot Inspection. J. Chem. Inf. Model. 2012, 52, 2044–2058. [Google Scholar] [CrossRef]







| Parameter | RF-CoMFA | RF-CoMFA (Test Set 16) |
|---|---|---|
| q2 | 0.527 | 0.751 |
| ONC | 6 | 4 |
| SEP | 0.694 | 0.395 |
| r2 | 0.905 | 0.911 |
| SEE | 0.258 | 0.236 |
| F-value | 73.716 | 53.689 |
| Q2 | - | 0.568 |
| BS-r2 | - | 0.932 |
| BS-SD | - | 0.038 |
| r2pred | - | 0.808 |
| LOF | - | 0.751 |
| rm2 | - | 0.523 |
| Δ rm2 | - | 0.120 |
 | ||||
| Compound | R1 | R2 | R3 | Predicted pIC50 |
| D01 |  |  |  | 8.20 |
| D02 |  | 8.236 | ||
| D03 |  | 8.612 | ||
| D04 |  |  | 8.50 | |
| D05 |  | 8.289 | ||
| D06 |  | 8.02 | ||
| D07 |  | 8.32 | ||
| D08 |  | 8.30 | ||
| Compound | Properties | Absorption (in %) | Distribution (in %) | Metabolism (in %) | Elimination (Liver Microsomal Stability) (in %) | Toxicity (in %) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPSA (in %) | AlogP | Passive Absorption (Permeability) | Blood-Brain Barrier Penetration | P-gp Substrates | CYP1A2 Inhibition | CYP2D6 Inhibition | CYP2C9 Inhibition | CYP2C19 Inhibition | CYP3A4 Inhibition | Human | Mouse | Rat | hERG Inhibition | |
| D1 | 111.80 | 3.36 | 52 | 67 | 39 | 55 | 34.50 | 57 | 49 | 44 | 53.38 | 77 | 66 | 48.43 |
| D2 | 97.97 | 2.96 | 55 | 65.50 | 43 | 53 | 39 | 57.20 | 57 | 47 | 51.48 | 76 | 61 | 54.33 |
| D3 | 125.44 | 3.30 | 46 | 57 | 42 | 56 | 32 | 49.60 | 44.67 | 48 | 54.28 | 71 | 59 | 51.28 |
| D4 | 106.51 | 1.33 | 55 | 65 | 28 | 53.50 | 22.33 | 56.10 | 47.08 | 41 | 54.46 | 76 | 63 | 43.17 |
| D5 | 143.81 | 1.04 | 44 | 68 | 45 | 45.50 | 19.50 | 56.90 | 50.33 | 43 | 53.19 | 79 | 69 | 41.17 |
| D6 | 106.51 | 1.85 | 58 | 61 | 37 | 50.50 | 23.33 | 55.20 | 49.89 | 44 | 50.63 | 78 | 52 | 43.63 |
| D7 | 106.51 | 3.27 | 55 | 59 | 41 | 45 | 27.33 | 54.80 | 54.33 | 49 | 54.21 | 73 | 56 | 46.30 |
| D8 | 106.51 | 3.99 | 50 | 56 | 38 | 46 | 29.17 | 55.40 | 48.67 | 43 | 48 | 78 | 63 | 50.30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhujbal, S.P.; He, W.; Hah, J.-M. Design of Novel IRAK4 Inhibitors Using Molecular Docking, Dynamics Simulation and 3D-QSAR Studies. Molecules 2022, 27, 6307. https://doi.org/10.3390/molecules27196307
Bhujbal SP, He W, Hah J-M. Design of Novel IRAK4 Inhibitors Using Molecular Docking, Dynamics Simulation and 3D-QSAR Studies. Molecules. 2022; 27(19):6307. https://doi.org/10.3390/molecules27196307
Chicago/Turabian StyleBhujbal, Swapnil P., Weijie He, and Jung-Mi Hah. 2022. "Design of Novel IRAK4 Inhibitors Using Molecular Docking, Dynamics Simulation and 3D-QSAR Studies" Molecules 27, no. 19: 6307. https://doi.org/10.3390/molecules27196307