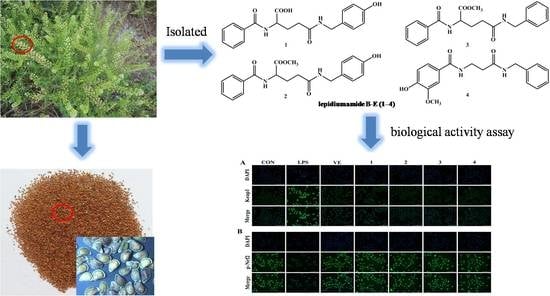
Four New Benzoylamide Derivatives Isolated from the Seeds of Lepidium apetalum Willd. and Ameliorated LPS-Induced NRK52e Cells via Nrf2/Keap1 Pathway
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Compounds
2.2. Effects of Compounds 1–4 on LPS-Induced NRK-52e Cell Viability
2.3. Effects of Compounds 1–4 on the Levels of Keap1 and p-Nrf2 in NRK-52e Cell Induced by LPS
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Materials
3.3. Extraction and Isolation
3.4. Compound Characterization
3.5. Cell Culture and MTT Assay
3.6. Immunofluorescence
3.7. Statistical Analysis
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kim, S.B.; Seo, Y.S.; Kim, H.S.; Lee, A.Y.; Chun, J.M.; Moon, B.C.; Kwon, B.I. Anti-asthmatic effects of lepidiiseuDescurainiae Semen plant species in ovalbumin-induced asthmatic mice. J. Ethnopharmacol. 2019, 244, 112083. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.D.; Tang, L.Y.; Zhou, G.H.; Kou, Z.Z.; Wang, T.; Wang, Z.J. Advances on Lepidii Semen and Descurainiae Semen. Chin. J. Chin. Mater. Med. 2014, 39, 4699–4708. [Google Scholar]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China MedicalScience and Technology Press: Beijing, China, 2020; Volume 1, p. 348. [Google Scholar]
- Nimrouzi, M.; Zarshenas, M.M. Phytochemical and pharmacological aspects of Descurainiasophia Webb ex Prantl: Modern and traditional applications. Avicenna. J.Phytomed. 2016, 6, 266–272. [Google Scholar] [PubMed]
- Gong, J.H.; Zhang, Y.L.; He, J.L.; Zheng, X.K.; Feng, W.S.; Wang, X.L.; Kuang, H.X.; Li, C.G.; Cao, Y.G. Extractions of oil from Descurainiasophia seed using supercritical CO2, chemical compositions by GC-MS and evaluation of the anti-tussive, expectorant and anti-asthmatic activities. Molecules 2015, 20, 13296–13312. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.S.; Li, C.G.; Zheng, X.K.; Li, L.L.; Chen, W.J.; Zhang, Y.L.; Cao, Y.G.; Gong, J.H.; Kuang, H.X. Three new sulphur glycosides from the seeds of Descurainiasophia. Nat. Prod. Res. 2016, 30, 1675–1681. [Google Scholar] [CrossRef] [PubMed]
- Choopani, R.; Ghourchian, A.; Hajimehdipoor, H.; Kamalinejad, M.; Ghourchian, F. Effect of Descurainiasophia (L.) Webb ex Prantl on adult functional constipation: Aprospective pilot study. J.Evid. Based. Complementary.Altern. Med. 2017, 22, 646–651. [Google Scholar] [CrossRef] [Green Version]
- Meng, Z.; Qiao, L.; Wang, Z.; Ma, Z.; Jia, J.; Chang, Q.; Li, W. AnalysisoftheconstituentsinSemenDescurainiaebyUPLC/Q TOF MS/MS. J.Pharm. SCI-US. 2015, 24, 303–309. [Google Scholar]
- Wu, X.; Yang, Y.; Huang, D. Effect of aqueous extract of Lepidiumapetalum on dog’s left ventricular function. J. Chin. Med. Mat. 1998, 21, 243–245. [Google Scholar]
- Shi, P.; Chao, L.; Wang, T.; Liu, E.; Han, L.; Zong, Q.; Li, X.; Zhang, Y.; Wang, T. New bioactive flavonoid glycosides isolated from the seeds of Lepidium apetalum Willd. Fitoterapia 2015, 130, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Shi, P.; Dong, Y.; Wang, T.; Li, X. New rare sinapoyl acylated flavonoid glycosides obtained from the seeds of Lepidium apetalumWilld. Molecules 2015, 20, 13982–13996. [Google Scholar] [CrossRef] [Green Version]
- Shi, P.; Li, X.; Zong, Q.; Han, L.; Wang, T.; Wang, Y. Flavonoid glycosides from seeds of Lepidium apetalum Willd II. J. Tropical. Subtropical. Bot. 2015, 23, 691–696. [Google Scholar]
- Shi, P.; Li, X.; Han, L.; Liu, E.; Wang, T.; Wang, Y. Isolation and identification of flavonoid glycosides from the seeds of Lepidium apetalum Willd. J Shenyang Pharm Univ. 2015, 32, 767–771. [Google Scholar]
- Li, M.; Zeng, M.; Zhang, Z.; Zhang, J.; Zhang, B.; Zhao, X.; Zheng, X.; Feng, W. Uridine derivatives from the seeds of Lepidium apetalum Willd. and their estrogenic effects. Phytochemistry 2018, 155, 45–52. [Google Scholar] [CrossRef]
- Li, M.; Zeng, M.; Zhang, Z.; Zhang, B.; Zhang, J.; Zheng, X.; Feng, W. Lepidiumuridine A: A new natural uridine derivative as a phytoestrogen isolated from the seeds of Lepidium apetalum Willd. J.Evid. Based. Complementary Altern. 2018, 2018, 2813465. [Google Scholar] [CrossRef]
- Li, M.; Qi, Z.; Hao, Y.; Lv, C.; Jia, L.; Wang, J.; Lu, J. New adducts of iriflophene and flavonoids isolated from sedum aizoon l. with potential antitumor activity. Molecule 2017, 22, 1859. [Google Scholar] [CrossRef] [Green Version]
- Buchynskyy, A.; Gillespie, J.R.; Hulverson, M.A.; McQueen, J.; Creason, S.A.; Ranade, R.M.; Duster, N.A.; Gelb, M.H.; Buckner, F.S. Discovery of N-(2-aminoethyl)-N-benzyloxyphenyl benzamides: New potent Trypanosoma brucei inhibitors. Bioorg. Med. Chem. 2017, 25, 1571–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huong, T.T.; Dung, D.T.; Huan, N.V.; Cuong, L.V.; Hai, P.T.; Huong, L.T.; Kim, J.; Kim, Y.G.; Han, S.B.; Nam, N.H. Novel N-hydroxybenzamides incorporating 2-oxoindoline with unexpected potent histone deacetylase inhibitory effects and antitumor cytotoxicity. Bioorg. Chem. 2017, 71, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Gershon, H.; Rodin, R. Substituted 2-aminothiosulfuric acids derived from alpha-amino acids. J Med Chem. 1965, 8, 864–866. [Google Scholar] [CrossRef] [PubMed]
- Engstroem, K. Post-infectional metabolites from infected potato tubers. Phytochemistry 1998, 49, 1585–1587. [Google Scholar] [CrossRef]
- Luo, Y.; Su, C.; Ding, N.; Qi, B.; Jia, F.; Xu, X.; Liu, X.; Wang, J.; Wang, X.; Tu, P.; et al. Lignan glycosides from Urenalobata. Molecules 2019, 24, 2850. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.L.; Cao, Y.G.; Kan, Y.X.; Ren, Y.J.; Zeng, M.N.; Wang, M.N.; He, C.; Chen, X.; Zheng, X.K.; Feng, W.S. Two new eremophilane-type sesquiterpenes from the fresh roots of RehmanniaGlutinosa. Phytochem. Lett. 2020, 42, 125–128. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, Z.; Fu, Y.; Shan, H.; Cui, S.; Wu, J. Gsta3 regulates tgf-β1-induced renal interstitial fibrosis in nrk-52e cells as a component of the pi3k–keap1/nrf2 pathway. J. Int. Med. Res. 2019, 47, 5787–5801. [Google Scholar] [CrossRef] [PubMed]




| No. | 1 a | 2 a | 3 b | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | 135.1 | 135.0 | 133.7 | |||
| 2,6 | 7.85 (2H, d, 7.5) | 128.5 | 7.85 (2H, d, 7.5) | 128.5 | 7.88 (2H, d, 8.0) | 127.4 |
| 3,5 | 7.45 (2H, t, 7.5) | 129.5 | 7.45 (2H, t, 7.5) | 129.6 | 7.47 (2H, t, 7.5) | 128.2 |
| 4 | 7.52 (1H, m) | 132.9 | 7.54 (1H, m) | 133.0 | 7.54 (1H, m) | 131.5 |
| 7 | 170.3 | 170.3 | 166.5 | |||
| 8 | 8.80 (1H, d, 7.0) | |||||
| 9 | 4.59 (1H, m) | 54.0 | 4.59 (1H, m) | 54.2 | 4.59 (1H, m) | 52.4 |
| 10 | 2.31 (1H, m) 2.14 (1H, m) | 28.2 | 2.30 (1H, m) 2.14 (1H, m) | 28.0 | 2.40 (2H, m) | 26.3 |
| 11 | 2.40 (2H, m) | 33.5 | 2.40 (2H, m) | 33.3 | 2.31 (1H, m) 2.14 (1H, m) | 31.6 |
| 12 | 174.7 | 174.5 | 171.3 | |||
| 13 | 8.36 (1H, t, 6.0) | |||||
| 14 | 4.22 (2H, d, 4.0) | 43.8 | 4.22 (2H, d, 4.0) | 43.8 | 4.22 (2H, d, 4.0) | 42.0 |
| 15 | 175.0 | 173.8 | 172.5 | |||
| 1′ | 130.5 | 130.5 | 139.4 | |||
| 2′,6′ | 7.07 (2H, d, 8.5) | 130.0 | 7.07 (2H, d, 8.5) | 130.0 | 7.21 (2H, d, 8.5) | 127.1 |
| 3′,5′ | 6.67 (2H, d, 8.5) | 116.2 | 6.69 (2H, d, 8.5) | 116.2 | 7.28 (2H, d, 8.5) | 128.2 |
| 4′ | 157.7 | 157.7 | 7.21 (2H, d, 8.5) | 126.7 | ||
| OCH3 | 3.73 (3H, s) | 52.8 | 3.64 (3H, s) | 51.8 | ||
| No. | δH | δC | δH | δC | |
|---|---|---|---|---|---|
| 1 | 121.6 | 10 | 3.55 (2H, t, 8.0) | 31.1 | |
| 2 | 7.43 (1H, d, 2.0) | 112.7 | 11 | 167.2 | |
| 3 | 151.1 | 13 | 4.52 (2H, s) | 48.2 | |
| 4 | 147.2 | 1′ | 135.8 | ||
| 5 | 6.83 (1H, d, 8.5) | 115.0 | 2′,6′ | 7.32 (2H, m) | 127.7 |
| 6 | 7.41 (1H, dd, 2.0, 8.5) | 123.3 | 3′,5′ | 7.39 (2H, m) | 128.7 |
| 7 | 163.9 | 4′ | 7.32 (1H, m) | 128.0 | |
| 9 | 3.92 (2H, t, 8.0) | 49.8 | OCH3 | 3.79 (3H, s) | 55.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Zhang, B.; Zeng, M.; Zhang, J.; Zhang, Z.; Feng, W.; Zheng, X. Four New Benzoylamide Derivatives Isolated from the Seeds of Lepidium apetalum Willd. and Ameliorated LPS-Induced NRK52e Cells via Nrf2/Keap1 Pathway. Molecules 2022, 27, 722. https://doi.org/10.3390/molecules27030722
Li M, Zhang B, Zeng M, Zhang J, Zhang Z, Feng W, Zheng X. Four New Benzoylamide Derivatives Isolated from the Seeds of Lepidium apetalum Willd. and Ameliorated LPS-Induced NRK52e Cells via Nrf2/Keap1 Pathway. Molecules. 2022; 27(3):722. https://doi.org/10.3390/molecules27030722
Chicago/Turabian StyleLi, Meng, Beibei Zhang, Mengnan Zeng, Jingke Zhang, Zhiguang Zhang, Weisheng Feng, and Xiaoke Zheng. 2022. "Four New Benzoylamide Derivatives Isolated from the Seeds of Lepidium apetalum Willd. and Ameliorated LPS-Induced NRK52e Cells via Nrf2/Keap1 Pathway" Molecules 27, no. 3: 722. https://doi.org/10.3390/molecules27030722