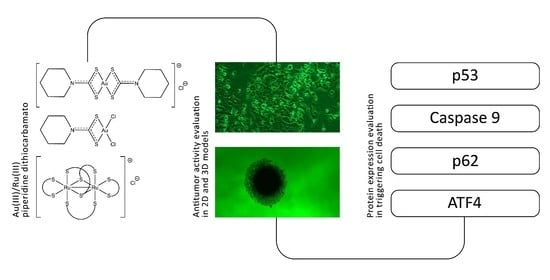
Gold(III) to Ruthenium(III) Metal Exchange in Dithiocarbamato Complexes Tunes Their Biological Mode of Action for Cytotoxicity in Cancer Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Syntheses and Characterization
2.2. Biological Activity
2.3. Investigation of the Mechanism of Action
3. Materials and Methods
- Synthesis of the K PipeDTC ligand. KOH (2.272 g, 40 mmol) was dissolved in 100 mL of ethanol and 4 mL (40.5 mmol) of piperidine was added with the solution mixture set at 0 °C. Successively, an excess of carbon disulfide (CS2) was added, leading to a yellow mixture, which was kept under vigorous stirring for 1 h. Afterwards, the solvent volume was reduced and the product precipitated by adding cold diethyl ether, filtered and washed with cold ethanol, before being dried under vacuum in the presence of P2O5. Piperidine dithiocarbamate potassium salt: aspect: white solid. Yield: 95%. Anal. Calc. C6H10KNS2 (MW = 199.38 g/mol): C, 36.14; H, 5.06; N, 7.03; S, 32.16. Found: C, 35.84; H, 5.02; N, 7.05; S, 32.22. 1H-NMR (CD3CN, 300.13 MHz): δ (ppm): 1.50 (m, 4H); 1.60 (m, 2H); 4.34 (t, 4H). Medium FT-IR (KBr): (cm−1) = 1417.12 (v N-CSS), 965.00 (va CSS).
- Synthesis of the precursor [AuI(pipeDTC)]2. H[AuCl4]∙xH2O (714.7 mg, 1.78 mmol) was dissolved in a saturated saline solution at 0 °C, followed by reduction Au(III)→Au(I) with 1 equivalent of Na2SO3 under stirring. The bright yellow solution turns colorless, and immediately 355.1 mg (1.78 mmol) of K PipeDTC was added, leading to the precipitation of an orange solid. The mixture was left under vigorous stirring for 5 min, and then the precipitate was centrifuged, washed three times with 5 mL of water and dried under vacuum in the presence of P2O5. Bis(piperidine dithiocarbamate)digold(I): aspect: green–yellow solid. Yield: 98%. Anal. Calc. C12H20Au2N2S4 (MW = 714.49 g/mol): C, 20.17; H, 2.82; N, 3.92; S, 17.95. Found: C, 20.20; H, 2.82; N, 3.99; S, 17.89. Medium FT-IR (KBr): (cm−1) = 1427.69 (v N-CSS), 967.97 (va CSS).
- Synthesis of [AuCl2(pipeDTC)]. [AuI(pipeDTC)]2] (0.5 mmol) was suspended in 50 mL of dichloromethane and the mixture was refluxed under stirring. After 10 min, an excess of Cl2 (generated in a separate flask by mixing 300 mg of MnO2 with 5 mL of concentrated HCl) was gurgled in the Au(I) suspension for 1 h. It was possible to observe that, after some minutes, the solution turned brown with the dissolution of the Au(I) precursor, and then became yellow. Then, the solution was cooled to room temperature and the solvent half-reduced. The compound was precipitated with diethyl ether, washed with 2 × 4 mL of diethyl ether and 2 × 5 mL of deionized water, and the orange product was dried under vacuum in the presence of P2O5. Dichloro(piperidine dithiocarbamate)gold(III): aspect: dark-yellow solid. Yield: 88%. Anal. Calc. C6H10AuCl2NS2 (MW = 428.15 g/mol): C, 16.83; H, 2.35; N, 3.27; S, 14.98. Found: C, 17.02; H, 2.40; N, 3.14; S, 15.02. 1H-NMR (CD3CN, 400.13 MHz): δ (ppm): 1.78 (s, 6H); 3.75 (s, 4H). Medium FT-IR (KBr): (cm−1) = 1582 (v N-CSS), 1006 (va CSS).
- Synthesis of [Au(pipeDTC)2]Cl. [AuIIICl2(pipeDTC)](123.1 mg, 0.288 mmol) was dissolved in CHCl3 (60 mL), and, in a separate flask, the ligand K pipeDTC (57.3 mg, 0.288 mmol) was dissolved in ethanol (1 mL). The ethanolic solution of the ligand was added dropwise to the Au(III) precursor, and the solution turned from yellow to light orange. After 10 min, the solvent was removed under reduced pressure. The residual solid was taken up with CHCl3, and KCl was removed by filtration. The final product was collected by precipitation with n-pentane and dried under vacuum in the presence of P2O5. Bis(piperidine dithiocarbamate)gold(III) chloride: aspect: yellow–orange solid. Yield: 82%. Anal. Calc. C12H20AuN2S4Cl (MW = 552.98 g/mol): C, 26.04; H, 3.61; N, 5.06; S, 23.19. Found: C, 25.86; H, 3.93; N, 4.83; S, 23,32. 1H-NMR (CD3CN, 400.13 MHz): δ (ppm): 1.79 (m12H); 3.80 (m, 8H). Medium FT-IR (KBr): (cm−1) = 1560 (v N-CSS), 1003 ( va CSS).
- Syntheses of [Ru2(pipeDTC)5]Cl and [Ru(pipeDTC)3]. Ruthenium(III)chloride trihydrate (818.9 mg, 3.13 mmol) and K pipeDTC (187.3 mg, 9.39 mmol) were separately dissolved in 5 mL of deionized water. The solution of ligand was added to Ru(III), and stirred at room temperature for 1 h, yielding a brown precipitate that was centrifuged, washed with 2 × 10 mL of deionized water and dried under vacuum in the presence of P2O5. After 24 h, the isolated product was re-dissolved in 20 mL of CH2Cl2 and the solution filtered to remove Ru oxides by-products. The obtained solution was purified via silica gel chromatography. A gradient from CH2Cl2 100% to CH2Cl2/MeOH 90%:10% was used to first elute the dark-green mononuclear complex [Ru(pipeDTC)3] and then the brown dinuclear derivative [Ru2(pipeDTC)5]Cl as a mixture of α and β isomers. Successively, the mixture of α, β-[Ru2(pipeDTC)5]Cl was isomerized to the thermodynamically stable β-[Ru2(pipeDTC)5]Cl by reflux in methanol for 8 h. Both the mononuclear and the β-dinuclear complexes were re-precipitated from CH2Cl2 diethyl ether, washed with n-pentane, and dried in vacuum in the presence of P2O5. β-pentakis(piperidine dithiocarbamate)diruthenium(III)chloride: aspect: brown solid. Yield: 30%. Anal. Calc. C30H50Ru2ClN5S10 (MW = 1039.00 g/mol): C, 34.64; H, 4.81; N, 6.73; S, 30.85. Found: C, 33.01; H, 4.73; N, 6.50; S, 30.37. 1H-NMR (CD3CN, 400.13 MHz): δ (ppm): 1.75 (m, 10H); 3.88 (m, 6H). Medium FT-IR (KBr): (cm−1) = 1505, 1441 (v N-CSS), 1001 (va CSS). Tris(piperidine dirhiocarbamate)ruthenium(III): aspect: dark-green solid. Yield: 34%. Anal. Calc. C18H30RuN3S6 (MW = 581.91 g/mol): C, 37.13; H, 5.16; N,7.22; S, 33.07. Found: C, 37.19; H, 5.01; N, 7.00; S, 33.97.1H-NMR (CD3CN, 400.13 MHz): δ (ppm): 26.02 (m, 6H); 21.95 (m, 6H); 1.01 (m, 12H); 3.95 (m, 6H). Medium FT-IR (KBr): (cm−1) 1488, 1455, 1440 (v N-CSS), 1002 (va CSS).
3.1. Cell Lines and Culture Conditions
3.2. Instrumentation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Cancer. Available online: https://www.who.int/health-topics/cancer#tab=tab_1 (accessed on 12 April 2021).
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal complexes in cancer therapy-an update from drug design perspective. Drug Des. Devel. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef] [Green Version]
- Cepeda, V.; Fuertes, M.; Castilla, J.; Alonso, C.; Quevedo, C.; Perez, J. Biochemical Mechanisms of Cisplatin Cytotoxicity. Anticancer. Agents Med. Chem. 2008, 7, 3–18. [Google Scholar] [CrossRef]
- Kelland, L.R. Preclinical perspectives on platinum resistance. Drugs 2000, 59, 1–8. [Google Scholar] [CrossRef]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef] [Green Version]
- Oun, R.; Wheate, N. Encyclopedia of Metalloproteins; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 9781461415336. [Google Scholar]
- Dilruba, S.; Kalayda, G.V. Platinum-based drugs: Past, present and future. Cancer Chemother. Pharmacol. 2016, 77, 1103–1124. [Google Scholar] [CrossRef]
- Reddy, T.S.; Privérprivér, S.H.; Mirzadeh, N.; Luwor, R.B.; Reddy, V.G.; Ramesan, S.; Bhargava, S.K. Antitumor and Antiangiogenic Properties of Gold(III) Complexes Containing Cycloaurated Triphenylphosphine Sulfide Ligands. Inorg. Chem. 2020. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, C.Y.; Nam, T.-G. Ruthenium Complexes as Anticancer Agents: A Brief History and Perspectives. Drug Des. Dev. Ther. 2020. [Google Scholar] [CrossRef]
- Lazarevi, T.; Rilak, A.; Bugar, Z.D. Platinum, palladium, gold and ruthenium complexes as anticancer agents: Current clinical uses, cytotoxicity studies and future perspectives. Eur. J. Med. Chem. 2017. [Google Scholar] [CrossRef]
- Fricker, S.P. Medical uses of gold compounds: Past, present and future. Gold Bull. 1996, 29, 53–60. [Google Scholar] [CrossRef] [Green Version]
- Notaro, A.; Gasser, G. Monomeric and dimeric coordinatively saturated and substitutionally inert Ru(II) polypyridyl complexes as anticancer drug candidates. Chem. Soc. Rev. 2017, 46, 7317–7337. [Google Scholar] [CrossRef] [PubMed]
- Graf, N.; Lippard, S.J. Redox activation of metal-based prodrugs as a strategy for drug delivery. Adv. Drug Deliv. Rev. 2012, 64, 993–1004. [Google Scholar] [CrossRef] [Green Version]
- Nardon, C.; Brustolin, L.; Fregona, D. Is matching ruthenium with dithiocarbamato ligands a potent chemotherapeutic weapon in oncology? Future Med. Chem. 2016, 8, 211–226. [Google Scholar] [CrossRef]
- Allardyce, C.S.; Dyson, P.J. Ruthenium in medicine: Current clinical uses and future prospects. Platin. Met. Rev. 2001, 45, 62–69. [Google Scholar]
- Kratz, F.; Messori, L. Spectral characterization of ruthenium(III) transferrin. J. Inorg. Biochem. 1993, 49, 79–82. [Google Scholar] [CrossRef]
- Giovagnini, L.; Sitran, S.; Castagliuolo, I.; Brun, P.; Corsini, M.; Zanello, P.; Zoleo, A.; Maniero, A.; Biondi, B.; Fregona, D. Ru(iii)-based compounds with sulfur donor ligands: Synthesis, characterization, electrochemical behaviour and anticancer activity. Dalt. Trans. 2008, 6699–6708. [Google Scholar] [CrossRef]
- Ronconi, L.; Fregona, D. The Midas touch in cancer chemotherapy: From platinum- to gold-dithiocarbamato complexes. Dalt. Trans. 2009, 10670–10680. [Google Scholar] [CrossRef]
- Borch, R.F.; Pleasants, M.E. Inhibition of cis-platinum nephrotoxicity by diethyldithiocarbamate rescue in a rat model. Proc. Natl. Acad. Sci. USA 1979, 76, 6611–6614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nardon, C.; Fregona, D. Gold(III) Complexes in the Oncological Preclinical Arena: From Aminoderivatives to Peptidomimetics. Curr. Top. Med. Chem. 2015, 16, 360–380. [Google Scholar] [CrossRef] [PubMed]
- Stoytcheva, M. Pesticides in the Modern World-Pesticides Use and Management; InTech: Radom, Poland, 2012. [Google Scholar]
- Hogarth, G. Metal-dithiocarbamate complexes: Chemistry and biological activity. Mini-Reviews Med. Chem. 2012, 12, 1202–1215. [Google Scholar] [CrossRef]
- Shi, Y.; Chu, W.; Wang, Y.; Wang, S.; Du, J.; Zhang, J.; Li, S.; Zhou, G.; Qin, X.; Zhang, C. Synthesis, characterization and cytotoxicity of the Au(III) complexes with cyclic amine-based dithiocarbamate ligands. Inorg. Chem. Commun. 2013, 30, 178–181. [Google Scholar] [CrossRef]
- Bonati, F.; Ugo, R. Organotin(IV) N,N-disubstituted dithiocarbamates. J. Organomet. Chem. 1967, 10, 257–268. [Google Scholar] [CrossRef]
- Nagy, E.M.; Pettenuzzo, A.; Boscutti, G.; Marchiò, L.; Dalla Via, L.; Fregona, D. Ruthenium(II/III)-based compounds with encouraging antiproliferative activity against non-small-cell lung cancer. Chem.-A Eur. J. 2012, 18, 14464–14472. [Google Scholar] [CrossRef]
- Boscutti, G.; Marchiò, L.; Ronconi, L.; Fregona, D. Insights into the reactivity of gold-dithiocarbamato anticancer agents toward model biomolecules by using multinuclear NMR spectroscopy. Chem.-A Eur. J. 2013, 19, 13428–13436. [Google Scholar] [CrossRef]
- Carcas, L. Gastric cancer review. J. Carcinog. 2014, 13, 14. [Google Scholar] [CrossRef]
- Worldwide Cancer Data | World Cancer Research Fund. Available online: https://www.wcrf.org/dietandcancer/cancer-trends/worldwide-cancer-data (accessed on 12 April 2021).
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical Cancer Models in Tumor Biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Nardon, C.; Boscutti, G.; Gabbiani, C.; Massai, L.; Pettenuzzo, N.; Fassina, A.; Messori, L.; Fregona, D. Cell and Cell-Free Mechanistic Studies on Two Gold(III) Complexes with Proven Antitumor Properties. Eur. J. Inorg. Chem. 2017, 2017, 1737–1744. [Google Scholar] [CrossRef] [Green Version]
- Bergamo, A.; Gaiddon, C.; Schellens, J.H.M.; Beijnen, J.H.; Sava, G. Approaching tumour therapy beyond platinum drugs: Status of the art and perspectives of ruthenium drug candidates. J. Inorg. Biochem. 2012, 106, 90–99. [Google Scholar] [CrossRef]
- Pan, Y.; Karns, K.; Herr, A.E. Microfluidic electrophoretic mobility shift assays for quantitative biochemical analysis. Electrophoresis 2014, 35, 2078–2090. [Google Scholar] [CrossRef] [Green Version]
- Klajner, M.; Licona, C.; Fetzer, L.; Hebraud, P.; Mellitzer, G.; Pfeffer, M.; Harlepp, S.; Gaiddon, C. Subcellular localization and transport kinetics of ruthenium organometallic anticancer compounds in living cells: A dose-dependent role for amino acid and iron transporters. Inorg. Chem. 2014, 53, 5150–5158. [Google Scholar] [CrossRef]
- Brustolin, L.; Pettenuzzo, N.; Nardon, C.; Quarta, S.; Montagner, I.; Pontisso, P.; Rosato, A.; Conte, P.; Merigliano, S.; Fregona, D. Labelled micelles for the delivery of cytotoxic Cu(II) and Ru(III) compounds in the treatment of aggressive orphan cancers: Design and biological in vitro data. J. Inorg. Biochem. 2020, 213, 111259. [Google Scholar] [CrossRef]
- Younger, S.T.; Rinn, J.L. P53 regulates enhancer accessibility and activity in response to DNA damage. Nucleic Acids Res. 2017, 45, 9889–9900. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of p53 in cell death and human cancers. Cancers 2011, 3, 994. [Google Scholar] [CrossRef]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanchet, A.; Bourgmayer, A.; Kurtz, J.E.; Mellitzer, G.; Gaiddon, C. Isoforms of the p53 family and gastric cancer: A ménage à trois for an unfinished affair. Cancers 2021, 13, 916. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Q.; Mahmood, T.; Yang, P.C. Western blot: Technique, theory and trouble shooting. N. Am. J. Med. Sci. 2014, 6, 160. [Google Scholar] [CrossRef] [PubMed]
- Chow, M.J.; Babak, M.V.; Wong, D.Y.Q.; Pastorin, G.; Gaiddon, C.; Ang, W.H. Structural determinants of p53-independence in anticancer ruthenium-arene Schiff-base complexes. Mol. Pharm. 2016, 13, 2543–2554. [Google Scholar] [CrossRef]
- Licona, C.; Spaety, M.E.; Capuozzo, A.; Ali, M.; Santamaria, R.; Armant, O.; Delalande, F.; Van Dorsselaer, A.; Cianferani, S.; Spencer, J.; et al. A ruthenium anticancer compound interacts with histones and impacts differently on epigenetic and death pathways compared to cisplatin. Oncotarget 2017, 8, 2568–2584. [Google Scholar] [CrossRef] [Green Version]
- Fisher, D.E. Apoptosis in cancer therapy: Crossing the threshold. Cell 1994, 78, 539–542. [Google Scholar] [CrossRef]
- Chow, M.J.; Alfiean, M.; Pastorin, G.; Gaiddon, C.; Ang, W.H. Apoptosis-independent organoruthenium anticancer complexes that overcome multidrug resistance: Self-assembly and phenotypic screening strategies. Chem. Sci. 2017, 8, 3641–3649. [Google Scholar] [CrossRef] [Green Version]
- O’Donovan, N.; Crown, J.; Stunell, H.; Hill, A.D.K.; McDermott, E.; O’Higgins, N.; Duffy, M.J. Caspase 3 in breast cancer. Clin. Cancer Res. 2003, 9, 738–742. [Google Scholar]
- Törmänen-Näpänkangas, U.; Soini, Y.; Kahlos, K.; Kinnula, V.; Pääkkö, P. Expression of caspases-3, -6 and -8 and their relation to apoptosis in non-small cell lung carcinoma. Int. J. Cancer 2001, 93, 192–198. [Google Scholar] [CrossRef]
- Shintani, M.; Osaw, K. Role of Autophagy in Cancer. Nat. Rev. Cancer. 2013, 7, 961–967. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Hu, Y.; Sheng, R. Autophagy Regulators as Potential Cancer Therapeutic agents: A Review. Curr. Top. Med. Chem. 2015, 15, 720–744. [Google Scholar] [CrossRef]
- Walton, M.I.; Wilson, S.C.; Hardcastle, I.R.; Mirza, A.R.; Workman, P. An evaluation of the ability of pifithrin-α and -β to inhibit p53 function in two wild type p53 human tumor cell lines. Mol. Cancer Ther. 2005, 4, 1369–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Zhou, Y.F.; Gong, J.Y.; Gao, C.B.; Li, S.L. Expression of autophagy-related protein LC3B, p62, and cytoplasmic p53 in human retinoblastoma tissues. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3152–3160. [Google Scholar] [PubMed]
- Licona, C.; Delhorme, J.B.; Riegel, G.; Vidimar, V.; Cerón-Camacho, R.; Boff, B.; Venkatasamy, A.; Tomasetto, C.; Da Silva Figueiredo Celestino Gomes, P.; Rognan, D.; et al. Anticancer activity of ruthenium and osmium cyclometalated compounds: Identification of ABCB1 and EGFR as resistance mechanisms. Inorg. Chem. Front. 2020, 7, 678–688. [Google Scholar] [CrossRef]
- Chow, M.J.; Babak, M.V.; Tan, K.W.; Cheong, M.C.; Pastorin, G.; Gaiddon, C.; Ang, W.H. Induction of the Endoplasmic Reticulum Stress Pathway by Highly Cytotoxic Organoruthenium Schiff-Base Complexes. Mol. Pharm. 2018, 15, 3020–3031. [Google Scholar] [CrossRef] [PubMed]
- Velasco, G.; Verfaillie, T.; Salazar, M.; Agostinis, P. Linking ER stress to autophagy: Potential implications for cancer therapy. Int. J. Cell Biol. 2010, 2010. [Google Scholar] [CrossRef] [Green Version]
- Rozpedek, W.; Pytel, D.; Mucha, B.; Leszczynska, H.; Diehl, J.A.; Majsterek, I. The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr. Mol. Med. 2016, 16, 533–544. [Google Scholar] [CrossRef]
- Singleton, D.C.; Harris, A.L. Targeting the ATF4 pathway in cancer therapy. Expert Opin. Ther. Targets 2012, 16, 1189–1202. [Google Scholar] [CrossRef] [PubMed]
- Stockert, J.C.; Blázquez-Castro, A.; Cañete, M.; Horobin, R.W.; Villanueva, Á. MTT assay for cell viability: Intracellular localization of the formazan product is in lipid droplets. Acta Histochem. 2012, 114, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, A.A.; Dmitrenko, V.V. Pitfalls of the MTT assay: Direct and off-target effects of inhibitors can result in over/underestimation of cell viability. Gene 2015, 574, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Lee, S.H.; Dominguez, R. Regulation of actin cytoskeleton dynamics in cells. Mol. Cells 2010, 29, 311–325. [Google Scholar] [CrossRef]






| Compound | v(N-CSS) (cm−1) | v(CSS) (cm−1) |
|---|---|---|
| K pipeDTC | 1417 | 965 |
| [Au(pipeDTC)2]Cl | 1560 | 1003 |
| [AuCl2(pipeDTC)] | 1582 | 1006 |
| [Ru(pipeDTC)3] | 1488, 1455, 1440 | 1002 |
| [Ru2(pipeDTC)5]Cl | 1505, 1441 | 1001 |
| Compound | α-CH2 (ppm) | β-CH2 (ppm) | γ-CH2 (ppm) |
|---|---|---|---|
| piperidine | 2.68 | 1.40 | 1.40 |
| K pipeDTC | 4.34 | 1.50 | 1.60 |
| [AuCl2(pipeDTC)] | 3.75 | 1.78 | 1.78 |
| [Au(pipeDTC)2]Cl | 3.80 | 1.79 | 1.79 |
| [Ru(pipeDTC)3] | 26.02 21.95 | 1.01 | 3.95 |
| [Ru2(pipeDTC)5]Cl | 3.88 | 1.55 | 1.55 |
| 48 h | 72 h | |||
|---|---|---|---|---|
| Compound | HCT116 | AGS | HCT116 | AGS |
| Au 1:1 | 0.3 ± 0.2 | 0.9 ± 0.2 | 1.1 ± 0.7 | 0.4 ± 0.2 |
| Au 1:2 | 0.7 ± 0.5 | 0.3 ± 0.2 | 1.0 ± 0.2 | 0.4 ± 0.2 |
| Ru 2:5 | 0.28 ± 0.07 | 0.65 ± 0.08 | 0.25 ± 0.09 | 0.5 ± 0.1 |
| cis-Pt | 12 ± 2 | 21 ± 2 | 5.6 ± 0.6 | 8 ± 2 |
| Ru 1:3 | >70 | >70 | >70 | >70 |
| Protein | 1° Antibody | Dilution | 2° Antibody | Dilution | Substrate |
|---|---|---|---|---|---|
| caspase-3 | polyclonal rabbit (Cell Signaling) | 1:1000 | anti-rabbit (goat) (GE HealthCare) | 1:5000 | Luminata Forte |
| p62 | monoclonal mouse (Santa Cruz) | 1:1000 | anti-mouse (goat) (GE HealthCare) | 1:5000 | Luminata Crescendo |
| p53 | polyclonal rabbit (Santa Cruz) | 1:1000 | anti-rabbit (goat) (GE HealthCare) | 1:5000 | Luminata Forte |
| ATF4 | Polyclonal rat (BioLegend) | 1:500 | anti-rat (rabbit) (GE HealthCare) | 1:10,000 | Luminata Forte |
| actin-β | Monoclonal mouse (Chemicon) | 1:1000 | anti-mouse (goat) (GE HealthCare) | 1:10,000 | Luminata Crescendo |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalla Pozza, M.; Orvain, C.; Brustolin, L.; Pettenuzzo, N.; Nardon, C.; Gaiddon, C.; Fregona, D. Gold(III) to Ruthenium(III) Metal Exchange in Dithiocarbamato Complexes Tunes Their Biological Mode of Action for Cytotoxicity in Cancer Cells. Molecules 2021, 26, 4073. https://doi.org/10.3390/molecules26134073
Dalla Pozza M, Orvain C, Brustolin L, Pettenuzzo N, Nardon C, Gaiddon C, Fregona D. Gold(III) to Ruthenium(III) Metal Exchange in Dithiocarbamato Complexes Tunes Their Biological Mode of Action for Cytotoxicity in Cancer Cells. Molecules. 2021; 26(13):4073. https://doi.org/10.3390/molecules26134073
Chicago/Turabian StyleDalla Pozza, Maria, Christophe Orvain, Leonardo Brustolin, Nicolò Pettenuzzo, Chiara Nardon, Christian Gaiddon, and Dolores Fregona. 2021. "Gold(III) to Ruthenium(III) Metal Exchange in Dithiocarbamato Complexes Tunes Their Biological Mode of Action for Cytotoxicity in Cancer Cells" Molecules 26, no. 13: 4073. https://doi.org/10.3390/molecules26134073