Fungal Quorum-Sensing Molecules and Inhibitors with Potential Antifungal Activity: A Review
Abstract
:1. Fungal Quorum Sensing: An Over View
2. Quorum Sensing in Various Fungal Species
2.1. Candida Albicans
2.2. Debaryomyces Hansenii
2.3. Cryptococcus Neoformans
2.4. Saccharomyces Cerevisiae
2.5. Neurospora Crassa
2.6. Penicillium Species
2.7. Aspergillus Species
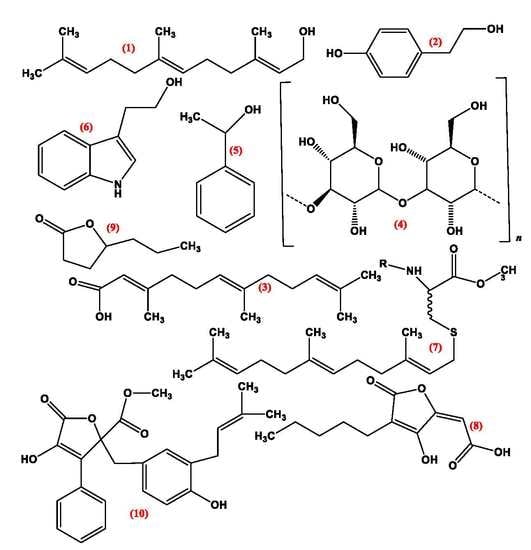
3. Quorum Sensing Molecules (QSMS)
3.1. Pheromones
3.2. Farnesol
3.3. Tyrosol
3.4. Volatile Organic Compounds
3.5. Lactone Containing Molecules
3.6. Lipids
4. Fungal Quorum Sensing Inhibitors (QSIs) with Potential Antifungal Activities
4.1. Farnesol
4.2. Others Fungal QSIs
4.3. The Antifungal Potential of QSMs/QSIs
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Padder, S.A.; Prasad, R.; Shah, A.H. Quorum sensing: A less known mode of communication among fungi. Microbiol. Res. 2018, 210, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Barriuso, J.; Hogan, D.A.; Keshavarz, T. Role of quorum sensing and chemical communication in fungal biotechnology and pathogenesis. FEMS Microbiol. 2018, 42, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Jangid, K. Book chapter Fungal Quorum Sensing Inhibitors. In Quorum Sensing vs Quorum Quenching: A Battle with no End in Sight; Kalia, V.C., Ed.; Springer: Mumbai, India, 2015. [Google Scholar]
- Srinivasan, R.; Devi, K.R.; Kannappan, A.; Pandian, S.K.; Ravi, A.V. Piper betle and its bioactive metabolite phytol mitigates quorum sensing mediated virulence factors and biofilm of nosocomial pathogen Serratia marcescens in vitro. J. Ethnopharmacol. 2016, 193, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Borea, L.; Naddeo, V.; Belgiorno, V.; Choo, K.H. Control of quorum sensing signals and emerging contaminants in electrochemical membrane bioreactors. Bioresour. Technol. 2018, 269, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Amache, R. Quorum Sensing for Improved Production of Industrially Useful Products from Filamentous Fungi. Ph.D. Thesis, University of Westminster, London, UK, 2014. Available online: http://www.westminster.ac.uk/research/westminsterresearch (accessed on 15 November 2018).
- Zhang, Y.P.; Li, J.Z.; Liu, F.Q.; Yan, H.; Li, J.L.; Zhang, X.; Jha, A.K. Specific quorum sensing signal molecules inducing the social behaviors of microbial populations in anaerobic digestion. Bioresour. Technol. 2019, 273, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.C.; Nielsen, L.K.; Vickers, C.E. Engineered quorum sensing using pheromone-mediated cell-to-cell communication in Saccharomyces cerevisiae. ACS Synth. Biol. 2013, 2, 136–149. [Google Scholar] [CrossRef]
- Yamagishi, J.F.; Saito, N.; Kaneko, K. Symbiotic Cell Differentiation and Cooperative Growth in Multicellular Aggregates. PLoS Comput. Biol. 2016, 12, e1005042. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, P.; Casadevall, A. Quorum sensing in fungi-a review. Med. Mycol. 2012, 50, 337–345. [Google Scholar] [CrossRef]
- Tong, Y.; Tang, J. Candida albicans infection and intestinal immunity. Microbiol. Res. 2017, 198, 27–35. [Google Scholar] [CrossRef]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef]
- Chen, H.; Fujita, M.; Feng, Q.; Clardy, J.; Fink, G.R. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. USA 2014, 101, 5048–5052. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, K.W.; Atkin, A.L.; Hornby, J.M. Quorum sensing in dimorphic fungi: Farnesol and beyond. Appl. Environ. Microbiol. 2016, 72, 3805–3813. [Google Scholar] [CrossRef]
- Lindsay, A.K.; Deveau, A.; Piispanen, A.E.; Hogan, D.A. Farnesol and cAMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot. Cell 2012, 11, 1219–1225. [Google Scholar] [CrossRef]
- Davis-Hanna, A.; Piispanen, A.E.; Stateva, L.I.; Hogan, D.A. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol. Microbiol. 2008, 67, 47–62. [Google Scholar] [CrossRef]
- Kebaara, B.W.; Langford, M.L.; Navarathna, D.H.; Dumitru, R.; Nickerson, K.W.; Atkin, A.L. Candida albicans Tup1 is involved in farnesol-mediated inhibition of filamentous-growth induction. Eukaryot. Cell 2008, 7, 980–987. [Google Scholar] [CrossRef]
- Hall, R.A.; Turner, K.J.; Chaloupka, J.; Cottier, F.; De Sordi, L.; Sanglard, D.; Levin, L.R.; Buck, J.; Mühlschlegel, F.A. The quorum sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans. Eukaryot. Cell 2011, 10, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Langford, M.L.; Hargarten, J.C.; Patefield, K.D.; Marta, E.; Blankenship, J.R.; Fanning, S. Candida albicans Czf1 and Efg1 coordinate the response to farnesol during quorum sensing, white-opaque thermal dimorphism, and cell death. Eukaryot. Cell 2013, 12, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, C.; Unoje, O.; Liu, H. Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proc. Natl. Acad. Sci. USA 2014, 111, 1975–1980. [Google Scholar] [CrossRef]
- Polke, M.; Sprenger, M.; Scherlach, K.; Albán-Proaño, M.C.; Martin, R.; Hertweck, C.; Jacobsen, I.D. A functional link between hyphal maintenance and quorum sensing in Candida albicans. Mol. Microbiol. 2017, 103, 595–617. [Google Scholar] [CrossRef]
- Johansen, P.; Jespersen, L. Impact of quorum sensing on the quality of fermented foods. Curr. Opin. Food Sci. 2017, 13, 16–25. [Google Scholar] [CrossRef]
- Cruz, J.M.; Dominguez, J.M.; Dominguez, H.; Parajo, J.C. Dimorphic behaviour of Debaryomyces hansenii grown on barley bran acid hydrolyzates. Biotechnol. Lett. 2000, 22, 605–610. [Google Scholar] [CrossRef]
- Gori, K.; Mortensen, H.D.; Arneborg, N.; Jespersen, L. Ammonia as a mediator for communication in strains of Debaryomyces hansenii and yeast species. J. Dairy Sci. 2007, 90, 5032–5041. [Google Scholar] [CrossRef]
- Armstrong-James, D.; Meintjes, G.; Brown, G.D. A neglected epidemic: Fungal infections in HIV/AIDS. Trends Microbiol. 2014, 22, 120–127. [Google Scholar] [CrossRef]
- Homer, C.M.; Summers, D.K.; Goranov, A.I.; Clarke, S.C.; Wiesner, D.L.; Diedrich, J.K.; Petnic, S. Intracellular action of a secreted peptide required for fungal virulence. Cell Host Microbe 2016, 19, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Chang, Y.C.; Nardone, G.; Kwon-Chung, K.J. TUP1 disruption in Cryptococcus neoformans uncovers a peptide-mediated density-dependent growth phenomenon that mimics quorum sensing. Mol. Microbiol. 2007, 64, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Chang, Y.C.; Varma, A.; Kwon-Chung, K.J. Regulatory diversity of TUP1 in Cryptococcus neoformans. Eukaryot. Cell 2009, 8, 1901–1908. [Google Scholar] [CrossRef] [PubMed]
- Madhani, H.D. Quorum sensing in fungi: Q&A. PLoS Pathog. 2011, 7, e1002301. [Google Scholar]
- Tian, X.Y.; He, G.J.; Hu, P.J.; Chen, L.; Tao, C.Y.; Cui, Y.L.; Shen, L.; Ke, W.X.; Xu, H.J. Cryptococcus neoformans sexual reproduction is controlled by a quorum sensing peptide. Nat. Microbiol. 2018, 3, 698–707. [Google Scholar] [CrossRef] [PubMed]
- May, R.C. Custom-Made quorum sensing for a eukaryote. Dev. Cell 2016, 37, 391–392. [Google Scholar] [CrossRef]
- Severin, F.F.; Meer, M.V.; Smirnova, E.A.; Knorre, D.A.; Skulachev, V.P. Natural causes of programmed death of yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2008, 1783, 1350–1353. [Google Scholar] [CrossRef]
- Wuster, A.; Babu, M.M. Transcriptional control of the quorum sensing response in yeast. Mol. BioSyst. 2009, 6, 134–141. [Google Scholar] [CrossRef]
- Avbelj, M.; Zupan, J.; Kranjc, L.; Raspor, P. Quorum-sensing kinetics in Saccharomyces cerevisiae: A symphony of ARO genes and aromatic alcohols. J. Agric. Food Chem. 2015, 63, 8544–8550. [Google Scholar] [CrossRef]
- Avbelj, M.; Zupan, J.; Raspor, P. Quorum-sensing in yeast and its potential in wine making. Appl. Microbiol. Biotechnol. 2016, 100, 7841–7852. [Google Scholar] [CrossRef] [PubMed]
- Hazelwood, L.A.; Daran, J.M.; Van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef]
- Ghosh, S.; Kebaara, B.W.; Atkin, A.L.; Nickerson, K.W. Regulation of aromatic alcohol production in Candida albicans. Appl. Environ. Microbiol. 2008, 74, 7211–7218. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fink, G.R. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006, 20, 1150–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, T. Glycosylphosphatidylinositol (GPI) Anchors: Biochemistry and Cell Biology: Introduction to a Thematic Review Series. J. Lipid Res. 2016, 57, 4–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebecca, C.D.; Hou, R.Y.; Matias, I.K.; Richard, C.G.; Bruno, F. The role of yeast ARO8, ARO9 and ARO10 genes in the biosynthesis of 3-(methylthio)-1-propanol from L-methionine during fermentation in synthetic grape medium. FEMS Yeast Res. 2018, 19. [Google Scholar] [CrossRef]
- Roca, M.G.; Arlt, J.; Jeffree, C.E.; Read, N.D. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell. 2005, 4, 911–919. [Google Scholar] [CrossRef]
- Raina, S.; Odell, M.; Keshavarz, T. Quorum sensing as a method for improving sclerotiorin production in Penicillium sclerotiorum. J. Biotechnol. 2010, 148, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ma, A.; Zhao, G. Effect of farnesol on Penicillium decumbens’s morphology and cellulase production. Bioresources 2011, 6, 3252–3259. [Google Scholar]
- Brown, S.H.; Zarnowski, R.; Sharpee, W.C.; Keller, N.P. Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus. Appl. Environ. Microbiol. 2008, 74, 5674–5685. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, F.; Roy, I.; Keshavarz, T. Impact of linoleic acid supplementation on lovastatin production in Aspergillus terreus cultures. App. Microbiol. Biotech. 2010, 88, 65–73. [Google Scholar] [CrossRef]
- Raina, S.; De Vizio, D.; Palonen, E.K. Is quorum sensing involved in lovastatin production in the filamentous fungus Aspergillus terreus? Process Biochem. 2012, 47, 843–852. [Google Scholar] [CrossRef]
- Affeldt, K.J.; Brodhagen, M.; Keller, N.P. Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein-coupled receptors. Toxins 2012, 4, 695–717. [Google Scholar] [CrossRef] [PubMed]
- Marwa, M.A.K.; Rasmey, A.H.M.; Zohri, A.A. The action mechanism and biocontrol potentiality of novel isolates of Saccharomyces cerevisiae against the aflatoxigenic Aspergillus flavus. Lett. Appl. Microbiol. 2018. [Google Scholar] [CrossRef]
- Williams, H.E.; Steele, J.C.; Clements, M.O. ɣ-Heptalactone is an endogenously produced quorum sensing molecule regulating growth and secondary metabolite production by Aspergillus nidulans. Appl. Microbiol. Biotech. 2012, 96, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Cottier, F.; Mühlschlegel, F.A. Communication in Fungi. Int. J. Microbiol. 2012, 351832. [Google Scholar] [CrossRef]
- Navarathna, D.H.; Hornby, J.M.; Krishnan, N.; Parkhurst, A.; Duhamel, G.E.; Nickerson, K.W. Effect of farnesol on a mouse model of systemic candidiasis, determined by use of a DPP3 knockout mutant of Candida albicans. Infect. Immun. 2007, 75, 1609–1618. [Google Scholar] [CrossRef]
- Kuranda, K.; Francois, J.; Palamarczyk, G. The isoprenoid pathway and transcriptional response to its inhibitors in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2010, 10, 14–27. [Google Scholar] [CrossRef]
- Asadollahi, M.A.; Maury, J.; Moller, K.; Nielsen, K.F.; Schalk, M.; Clark, A.; Nielsen, J. Production of plant sesquiterpenes in Saccharomyces cerevisiae: Effect of ERG9 repression on sesquiterpene biosynthesis. Biotechnol. Bioeng. 2008, 99, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Krom, B.P.; Levy, N.; Meijler, M.M.; Jabra-Rizk, M.A. Farnesol and Candida albicans: Quorum sensing or not quorum sensing? Isr. J. Chem. 2016, 56, 295–301. [Google Scholar] [CrossRef]
- Monteiro, D.R.; Feresin, L.P.; Arias, L.S.; Barão, V.A.R.; Barbosa, D.B.; Delbem, A.C.B. Effect of tyrosol on adhesion of Candida albicans and Candida glabrata to acrylic surfaces. Med. Mycol. 2015, 53, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Alem, M.A.; Oteef, M.D.; Flowers, T.H. Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot. Cell 2006, 5, 1770–1779. [Google Scholar] [CrossRef]
- Nickerson, K.W.; Atkin, A.L.; Hornyby, J.M. Quorum sensing in dimorphic fungi: Farnesol and beyond. Appl. Environ. Microbiol. 2006, 72, 3805–3813. [Google Scholar] [CrossRef]
- Chitarra, G.S.; Abee, T.; Rombouts, F.M.; Posthumus, M.A.; Dijksterhuis, J. Germination of Penicillium paneum Conidia is regulated by 1-octen-3-ol, a volatile self-inhibitor. Appl. Environ. Microbiol. 2004, 70, 2823–2829. [Google Scholar] [CrossRef] [PubMed]
- Chitarra, G.S.; Abee, T.; Rombouts, F.M.; Posthumus, M.A.; Dijksterhuis, J. 1-Octen-3-ol inhibits conidia germination of Penicillium paneum despite of mild effects on membrane permeability, respiration, intracellular pH, and changes the protein composition. FEMS Microbiol. Ecol. 2005, 54, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Morath, S.U.; Hung, R.; Bennett, J.W. Fungal volatile organic compounds: A review with emphasis on their biotechnological potential. Fungal Biol. Rev. 2012, 26, 73–83. [Google Scholar] [CrossRef]
- Palkova, Z.; Janderova, B.; Gabriel, J.; Zikanova, B.; Pospisek, M.; Forstova, J. Ammonia mediates communication between yeast colonies. Nature 1997, 390, 532–536. [Google Scholar] [CrossRef]
- Nemcovic, M.; Jakubíková, L.; Víden, I.; Farkas, V. Induction of conidiation by endogenous volatile compounds in Trichoderma spp. FEMS Microbiol. Lett. 2008, 284, 231–236. [Google Scholar]
- Schimmel, T.G.; Coffman, A.D.; Parsons, S.J. Effect of butyrolactone ion the producing fungus, Aspergillus terreus. Appl. Environ. Microbiol. 1998, 64, 3707–3712. [Google Scholar] [PubMed]
- Palonen, E.; Neffling, M.R.; Raina, S. Butyrolactone I quantification from lovastatin producing Aspergillus terreus using tandem mass spectrometry-evidence of signalling functions. Microorganisms 2014, 2, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Krzyczkowska, J.; Phan-Thi, H.; Waché, Y. Chapter, Lactone Formation in Yeast and Fungi. In Fungal Metabolites; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer: Berlin/Germany, Germany, 2017; pp. 461–498. [Google Scholar] [CrossRef]
- Palonen, E.K.; Raina, S.; Brandt, A.; Meriluoto, J.; Keshavarz, T.; Soini, J.T. Melanisation of Aspergillus terreus-Is Butyrolactone I Involved in the Regulation of Both DOPA and DHN Types of Pigments in Submerged Culture? Microorganisms 2017, 5, 22. [Google Scholar] [CrossRef]
- Palonen, E.K.; Raina, S.; Brandt, A.; Meriluoto, J.; Keshavarz, T.; Soini, J.T. Transcriptomic Complexity of Aspergillus terreus Velvet Gene Family under the Influence of Butyrolactone, I. Microorganisms 2017, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Gessler, N.N.; Filippovich, S.Y.; Bachurina, G.P.; Kharchenko, E.A.; Groza, N.V.; Belozerskaya, T.A. Oxylipins and oxylipin synthesis pathways in fungi. Appl. Biochem. Microbiol. 2017, 53, 628–639. [Google Scholar] [CrossRef]
- Tsitsigiannis, D.I.; Kowieski, T.M.; Zarnowski, R.; Keller, N.P. Conserved cross kingdom oxylipins modulate Aspergillus nidulans development, secondary metabolism and seed colonization. Arch. Microbiol. 2005, 151, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Tsitsigiannis, D.I.; Zarnowski, R.; Keller, N.P. The Lipid Body Protein, PpoA, Coordinates Sexual and Asexual Sporulation in Aspergillus Nidulans. J. Biol. Chem. 2004, 279, 11344–11353. [Google Scholar] [CrossRef]
- Sebolai, O.M.; Pohl, C.H.; Botes, P.J.; Strauss, C.J.; van Wyk, P.W.J.; Botha, A.; Kocka, J.L.F. 3-Hydroxy fatty acids found in capsules of Cryptococcus Neoformans. Can. J. Microbiol. 2007, 53, 809–812. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.P.; Kock, J.L.F.; Van Wyk, P.W.J. The distribution of oxylipins in the yeast dipodascopsis uninucleata as investigated by immunofluorescence and electron microscopy. Prostaglandins 1999, 59, 155–162. [Google Scholar] [CrossRef]
- Duan, L.K.L.; Pan, Q.H.; Wang, Y.Q.; Ye, Q.; Duan, C.Q.; Yan, G.L. Influence of addition of unsaturated fatty acids on fatty acid composition of Saccharomyces cerevisiae and wine aroma compounds. Sci. Agric. Sin. 2016, 49, 1960–1978. [Google Scholar] [CrossRef]
- Goodrich-Tanrikulu, M.; Howe, K.; Stafford, A.; Nelson, M.A. Novel Neurospora crassa mutants with altered synthesis of polyunsaturated fatty acids. Arch. Microbiol. 1998, 144, 1713–1720. [Google Scholar] [CrossRef]
- Yu, Y.D.; Amich, J.; Will, C.; Eagle, C.E.; Dyer, P.S.; Krappmann, S. The novel Aspergillus fumigatus MAT1-2-4 mating-type gene is required for mating and cleistothecia formation. Fungal Genet. Biol. 2017, 108, 1–12. [Google Scholar] [CrossRef]
- Das, U.N. Arachidonic acid and other unsaturated fatty acids and some of their metabolites function as endogenous antimicrobial molecules: A review. J. Adv. Res. 2018, 11, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Erb-Downward, J.R.; Noverr, M.C. Characterization of prostaglandin E2 production by Candida albicans. Infect. Immun. 2007, 75, 3498–3505. [Google Scholar] [CrossRef]
- Grozer, Z.; Toth, A.; Toth, R.; Kecskemeti, A.; Vagvolgyi, C.; Nosanchuk, J.D.; Szekeres, A.; Gacser, A. Candida parapsilosis produces prostaglandins from exogenous arachidonic acid and OLE2 is not required for their synthesis. Virulence 2015, 6, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.I. Toxigenic fungi and mycotoxins. Br. Med. Bull. 2000, 56, 184–192. [Google Scholar] [CrossRef] [Green Version]
- Cordeiro, R.A.; Nogueira, G.C.; Brilhante, R.S.N.; Teixeira, C.E.C.; Mourão, C.I.; Castelo-Branco, D. Farnesol inhibits in vitro growth of the Cryptococcus neoformans species complex with no significant communication in fungi. Int. J. Microbiol. 2012, 159, 375–380. [Google Scholar]
- Sardi, J.D.O.; Pitangui, N.D.; Rodriguez-Arellanes, G.; Taylor, M.L.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Highlights in pathogenic fungal biofilms. Rev. Iberoam. Micol. 2014, 31, 22–29. [Google Scholar] [CrossRef]
- Nunes, T.; Cardoso, P.; Freitas, R.; Figueira, E. Protective effects of farnesol on a Rhizobium strain exposed to cadmium. Ecotox. Environ. Saf. 2018, 165, 622–629. [Google Scholar] [CrossRef]
- Wongsuk, T.; Pumeesat, P.; Luplertlop, N. Fungal quorum sensing molecules: Role in fungal morphogenesis and pathogenicity. J. Basic Microbiol. 2016, 56, 440–447. [Google Scholar] [CrossRef]
- Adelaide, C.F.; Deize, E.A.; Mirlane, S.C.; Isabella, T.B.; Liliana, B.M.L.; Maristela, P.; Andre, C.A. Development, characterization, and in vitro-in vivo evaluation of polymeric nanoparticles containing miconazole and farnesol for treatment of vulvovaginal candidiasis. Med. Mycol. 2019, 57, 52–62. [Google Scholar] [CrossRef]
- Pammi, M.; Liang, R.; Hicks, J.; Barrish, J.; Versalovic, J. Farnesol decreases biofilms of Staphylococcus epidermidis and exhibits synergy with nafcillin and vancomycin. Pediatr. Res. 2011, 70, 578–583. [Google Scholar] [CrossRef]
- Gomes, F.I.A.; Teixeira, P.; Cerca, N.; Azeredo, J.; Oliveira, R. Effect of farnesol on structure and composition of Staphylococcus epidermidis biofilm matrix. Curr. Microbiol. 2011, 63, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Pumeesat, P.; Wongsuk, T.; Muangkaew, W.; Luplertlop, N. Growth-inhibitory effects of farnesol against Scedosporium boydii and Lomentospora prolificans. Southeast Asian J. Trop. Med. Public. Health 2017, 48, 170–178. [Google Scholar]
- Zhu, H.; Liu, W.; Tian, B.; Liu, H.; Ning, S. Inhibition of quorum sensing in the opportunistic pathogenic bacterium Chromobacterium violaceum by an extract from fruiting bodies of lingzhi or reishi medicinal mushroom Ganoderma lucidum. Int. J. Med. Mushroom 2011, 13, 559–564. [Google Scholar] [CrossRef]
- Ueda, M.; Kubo, T.M.K.; Nakamura, K. Purification and characterization of fibrinolytic alkaline protease from Fusarium sp. BLB. Appl. Microbiol. Biotechnol. 2007, 74, 331–338. [Google Scholar] [CrossRef]
- Wang, K.F.; Sui, K.Y.; Guo, C.; Liu, C.Z. Quorum sensing molecule-farnesol increased the production and biological activities of extracellular polysaccharide from Trametes versicolor. Int. J. Biol. Macromol. 2017, 104, 377–383. [Google Scholar] [CrossRef]
- Rasmussen, T.B.; Skindersoe, M.E.; Bjarnsholt, T.; Phipps, R.K.; Christensen, K.B.; Jensen, P.O. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiologys 2005, 151, 1325–1340. [Google Scholar] [CrossRef]
- Petrović, J.; Glamočlija, J.; Stojković, D.; Nikolić, M.; Ćirić, A. Bioactive composition, antimicrobial activities and the influence of Agrocybe aegerita (Brig.) Sing on certain quorum-sensing-regulated functions and biofilm formation by Pseudomonas aeruginosa. Food Funct. 2014, 5, 3296–3303. [Google Scholar] [CrossRef]
- Soković, M.; Ćirić, A.; Glamočlija, J.; Nikolić, M.; Griensven, L.J.L.D. Agaricus blazei hotwater extract shows anti quorumsensing activity in the nosocomial human pathogen Pseudomonas aeruginosa. Molecules 2014, 19, 4189–4199. [Google Scholar] [CrossRef]
- Fernandes, Â.; Petrović, J.; Stojković, D.; Barros, L.; Glamočlija, J.; Soković, M.; Martins, A.; Ferreira, C.F.R.I. Polyporus squamosus (Huds.) Fr from different origins: Chemical characterization, screening of the bioactive properties and specific antimicrobial effects against Pseudomonas aeruginosa. LWT-Food Sci. Technol. 2016, 69, 91–97. [Google Scholar] [CrossRef]
- Kostić, M.; Smiljković, M.; Petrović, J.; Glamočlija, J.; Barros, L.; Ferreira, C.F.R.I.; Ćirić, A.; Soković, M. Chemical, nutritive composition and wide-broad bioactive properties of honey mushroom Armillaria mellea. Food Funct. 2017, 8, 3239–3249. [Google Scholar] [CrossRef] [PubMed]
- Pattnaik, S.; Ahmed, T.; Ranganathan, S.K.; Ampasala, D.R.; Sarma, V.V.; Busi, S. Aspergillus ochraceopetaliformis SSP13 modulates quorum sensing regulated virulence and biofilm formation in Pseudomonas aeruginosa PAO1. Biofouling 2018, 34, 410–425. [Google Scholar] [CrossRef]
- Ikeh, M.; Ahmed, Y.; Quinn, J. Phosphate acquisition and virulence in human fungal pathogens. Microorganisms 2017, 5, 48. [Google Scholar] [CrossRef]
- Gonzalez, J.M.; Rodriguez, C.A.; Agudelo, M.; Zuluaga, A.F.; Vesga, O. Antifungal pharmacodynamics: Latin America’s perspective. Braz. J. Infect. Dis. 2017, 21, 79–87. [Google Scholar] [CrossRef]
- Su, H.; Han, L.; Ding, N.; Guan, P.; Hu, C.; Huang, X. Bafilomycin C1 exert antifungal effect through disturbing sterol biosynthesis in Candida albicans. J. Antibiot. 2018, 71, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Han, L.; Huang, X. Potential targets for the development of new antifungal drugs. J. Antibiot. 2018, 71, 978–991. [Google Scholar] [CrossRef]
- Sawant, B.; Khan, T. Recent advances in delivery of antifungal agents for therapeutic management of candidiasis. Biomed Pharmacother. 2017, 96, 1478–1490. [Google Scholar] [CrossRef]
- Rodrigues, M.E.; Silva, S.; Azeredo, J.; Henriques, M. Novel strategies to fight Candida species infection. Crit. Rev. Microbiol. 2016, 42, 594–606. [Google Scholar]
- Campoy, S.; Adrio, J.L. Antifungals. Biochem Pharmacol. 2017, 133, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Semighini, C.P.; Hornby, J.M.; Dumitru, R.; Nickerson, K.W.; Harris, S.D. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol. Microbiol. 2016, 59, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Fairn, G.D.; Macdonald, K.; McMaster, C.R. A chemogenomic screen in Saccharomyces cerevisiae uncovers a primary role for the mitochondria in farnesol toxicity and its regulation by the Pkc1 pathway. J. Biol. Chem. 2007, 282, 4868–4874. [Google Scholar] [CrossRef]
- Lorek, J.; Poggeler, S.; Weide, M.R.; Breves, R.; Bockmuhl, D.P. Influence of farnesol on the morphogenesis of Aspergillus niger. J. Basic Microbiol. 2008, 48, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Semighini, C.P.; Murray, N.; Harris, S.D. Inhibition of Fusarium graminearum growth and development by farnesol. FEMS Microbiol. Lett. 2008, 279, 259–264. [Google Scholar] [CrossRef]
- Derengowski, L.S.; De-Souza-Silva, C.; Braz, S.V.; MelloDe-Sousa, T.M. Antimicrobial effect of farnesol a Candida albicans quorum sensing molecule on Paracoccidioides brasiliensis growth and morphogenesis. Ann. Clin. Microbiol. Antimicrob. 2009, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, K.; Ebel, F.; Dirr, F.; Routier, F.H.; Heesemann, J.; Wagener, J. Farnesol misplaces tip-localized Rho proteins and inhibits cell wall integrity signalling in Aspergillus fumigatus. Mol. Microbiol. 2010, 76, 1191–1204. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Zhou, Y.; Wei, X. Farnesol induces apoptosis-like cell death in the pathogenic fungus Aspergillus flavus. Mycologia 2014, 106, 881–888. [Google Scholar] [CrossRef]
- Sharma, M.; Prasad, R. The quorum-sensing molecule farnesol is a modulator of drug efflux mediated by ABC multidrug transporters and synergizes with drugs in Candida albicans. Antimicrob. Agents Chemother. 2011, 55, 4834–4843. [Google Scholar] [CrossRef]
- Kim, D.; Sengupta, A.; Niepa, T.H.; Lee, B.H.; Weljie, A.; Freitas-Blanco, V.S.; Murata, R.M.; Stebe, K.J.; Lee, D.; Koo, H. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 2017, 7, 41332. [Google Scholar] [CrossRef]
- Hornby, J.M.; Jensen, E.C.; Lisec, A.D.; Tasto, J.J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef]
- Machida, K.; Tanaka, T.; Yano, Y.; Otani, S.; Taniguchi, M. Farnesol-induced growth inhibition in Saccharomyces cerevisiae by a cell cycle mechanism. Microbiology 1999, 145, 293–299. [Google Scholar] [CrossRef] [Green Version]
- Savoldi, M.; Malavazi, I.; Soriani, F.M.; Capellaro, J.L.; Kitamoto, K.; Da Ferreira, K.E.S.; Goldman, M.H.S.; Goldman, G.H. Farnesol induces the transcriptional accumulation of the Aspergillus nidulans apoptosis-inducing factor (AIF)-like mitochondrial oxidoreductase. Mol. Microbiol. 2008, 70, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, T.; Logue, M.E.; Reynolds, K.; Grenon, M.; Lowndes, N.F.; Butler, G. Transcriptional response of Candida parapsilosis following exposure to farnesol. Antimicrob. Agents Chemother. 2007, 51, 2304–2312. [Google Scholar] [CrossRef]
- Henriques, M.; Martins, M.; Azeredo, J.; Oliveira, R. Effect of farnesol on Candida dubliniensis morphogenesis. Lett. Appl. Microbiol. 2007, 44, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Kovács, R.; Bozó, A.; Gesztelyi, R.; Domán, M.; Kardos, G.; Nagy, F.; Tóth, Z.; Majoros, L. Effect of caspofungin and micafungin in combination with farnesol against Candida parapsilosis biofilms. Int. J. Antimicrob Agents. 2016, 47, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Dižová, S.; Bujdáková, H. Properties and role of the quorum sensing molecule farnesol in relation to the yeast Candida albicans. Pharmazie 2017, 72, 307–312. [Google Scholar]
- Xia, J.; Qian, F.; Xu, W.; Zhang, Z.; Wei, X. In vitro inhibitory effects of farnesol and interactions between farnesol and antifungals against biofilms of Candida albicans resistant strains. Biofouling 2017, 33, 283–293. [Google Scholar] [CrossRef]
- Chen, S.; Xia, J.; Li, C.; Zuo, L.; Wei, X. The possible molecular mechanisms of farnesol on the antifungal resistance of C. albicans biofilms: The regulation of CYR1 and PDE2. BMC Microbiol. 2018, 18, 203. [Google Scholar] [CrossRef]
- Arias, L.S.; Delbem, A.C.B.; Fernandes, R.A.; Barbosa, D.B.; Monteiro, D.R. Activity of tyrosol against single and mixed-species oral biofilms. J. Appl. Microbiol. 2016, 120, 1240–1249. [Google Scholar] [CrossRef] [Green Version]
- Do Vale, L.R.; Delbem, A.C.B.; Arias, L.S.; Fernandes, R.A.; Vieira, A.P.M.; Barbosa, D.B.; Monteiro, D.R. Differential effects of the combination of tyrosol with chlorhexidine gluconate on oral biofilms. Oral Dis. 2017, 23, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, D.R.; Arias, L.S.; Fernandes, R.A.; Deszo da Silva, L.F.; de Castilho, M.O.V.F.; da Rosa, T.O.; Delbem, A.C.B. Antifungal activity of tyrosol and farnesol used in combination against Candida species in the planktonic state or forming biofilms. J Appl. Microbiol. 2017, 123, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Brilhante, R.S.N.; Érica, P.C.; Rita, A.C.L.; Francisca, J.F.M. Terpinen-4-ol, tyrosol, and-lapachone as potential antifungals against dimorphic fungi. Braz. J. Microbiol. 2016, 4, 917–924. [Google Scholar]
- Rasamiravaka, T.; El Jaziri, M. Quorum-Sensing Mechanisms and Bacterial Response to Antibiotics in P. aeruginosa. Curr. Microbiol. 2016, 73, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Rico, M.; Olivares-Pacheco, J.; Alvarez-Ortega, C.; Camara, M.; Martinez, J.L. Role of the multidrug resistance efflux pump MexCD-OprJ in the Pseudomonas aeruginosa quorum sensing response. Front. Microbiol. 2018, 9, 2752. [Google Scholar] [CrossRef]
- Borges, C.A.; Maluta, R.P.; Beraldo, L.G.; Cardozo, M.V.; Guastalli, E.A.L.; Kariyawasam, S.; DebRoy, C.; Avila, F.A. Captive and free-living urban pigeons (Columba livia) from Brazil as carriers of multidrug-resistant pathogenic Escherichia coli. Vet. J. 2017, 219, 65–67. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehmood, A.; Liu, G.; Wang, X.; Meng, G.; Wang, C.; Liu, Y. Fungal Quorum-Sensing Molecules and Inhibitors with Potential Antifungal Activity: A Review. Molecules 2019, 24, 1950. https://doi.org/10.3390/molecules24101950
Mehmood A, Liu G, Wang X, Meng G, Wang C, Liu Y. Fungal Quorum-Sensing Molecules and Inhibitors with Potential Antifungal Activity: A Review. Molecules. 2019; 24(10):1950. https://doi.org/10.3390/molecules24101950
Chicago/Turabian StyleMehmood, Arshad, Guorong Liu, Xin Wang, Guannan Meng, Chengtao Wang, and Ya Liu. 2019. "Fungal Quorum-Sensing Molecules and Inhibitors with Potential Antifungal Activity: A Review" Molecules 24, no. 10: 1950. https://doi.org/10.3390/molecules24101950