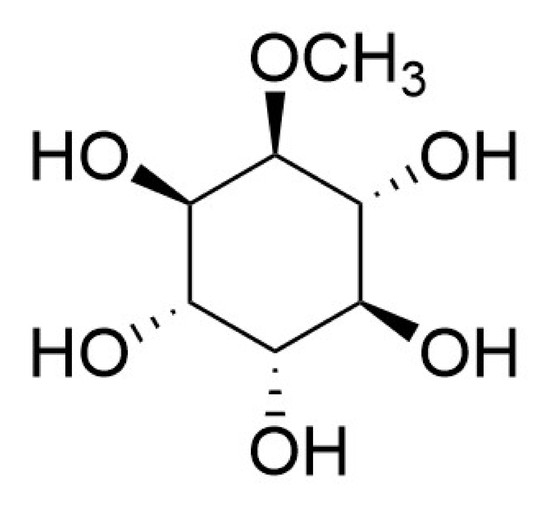
l-Quebrachitol Promotes the Proliferation, Differentiation, and Mineralization of MC3T3-E1 Cells: Involvement of the BMP-2/Runx2/MAPK/Wnt/β-Catenin Signaling Pathway
Abstract
:1. Introduction
2. Results
2.1. Extraction and Purification of l-quebrachitol from Rubber Latex Serum
2.2. l-quebrachitol Enhances the Cell Viability of Pre-Osteoblastic MC3T3-E1 Cells
2.3. l-quebrachitol Promotes Cell DNA Synthesis
2.4. l-quebrachitol Promotes Differentiation and Mineralization of Osteoblast Cells
2.5. l-quebrachitol Up-Regulates mRNA Expression of BMP-2, Runx2, and Osteogenesis Markers
2.6. l-quebrachitol Increases Expression of a Gene Involved in the MAPK and Wnt/β-Catenin Signaling Pathways
2.7. Western Blot Analysis of BMP-2 and Runx2 Expression
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Collection and Fractionation of Fresh Latex
4.3. Purification of l-quebrachitol
4.4. Cell Culture and Differentiation
4.5. Cytotoxicity Assay
4.6. Alkaline Phosphatase (ALP) Activity Assay
4.7. Alizarin Red S Staining
4.8. Cell Cycle Analysis
4.9. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.10. Western Blot Analysis
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hadjidakis, D.J.; Androulakis, I.I. Bone remodeling. Ann. N. Y. Acad. Sci. 2006, 1092, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Masi, L. Epidemiology of osteoporosis. Clin. Cases Miner. Bone MeTable 2008, 5, 11–13. [Google Scholar]
- An, J.; Yang, H.; Zhang, Q.; Liu, C.; Zhao, J.; Zhang, L.; Chen, B. Natural products for treatment of osteoporosis: The effects and mechanisms on promoting osteoblast-mediated bone formation. Life Sci. 2016, 147, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, R.T.; Ge, C.; Xiao, G.; Roca, H.; Jiang, D. Transcriptional regulation of osteoblasts. Ann. N. Y. Acad. Sci. 2007, 1116, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.D.; Yang, K.J.; Park, B.R.; Son, C.W.; Jang, H.J.; Ku, S.K. Antiosteoporotic effect of polycan, β-glucan from Aureobasidium, in ovariectomized osteoporotic mice. Nutrition 2007, 23, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Han, D.-W.; Park, B.J.; Baek, H.S.; Takatori, K.; Aihara, M.; Tsubaki, K.; Park, J.-C. The influences of β-glucan associated with BMP-7 on MC3T3-E1 proliferation and osteogenic differentiation. Key Eng. Mater. 2005, 288–289, 241–244. [Google Scholar] [CrossRef]
- Wittrant, Y.; Gorin, Y.; Woodruff, K.; Horn, D.; Abboud, H.E.; Mohan, S.; Abboud-Werner, S.L. High D(+)glucose concentration inhibits RANKL-induced osteoclastogenesis. Bone 2008, 42, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yue, F.; Wang, J.; Chen, L.; Qi, W. High glucose inhibits receptor activator of nuclear factor-κB ligand-induced osteoclast differentiation via downregulation of v-ATPase V0 sSubunit D2 and dendritic cell specific transmembrane protein. Mol. Med. Rep. 2015, 11, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Noguchi, C.; Kamitori, K.; Dong, Y.; Hirata, Y.; Hossain, M.A.; Tsukamoto, I.; Tokuda, M.; Yamaguchi, F. Rare sugar D-allose strongly induces thioredoxin-interacting protein and inhibits osteoclast differentiation in Raw264 Cells. Nutr. Res. 2012, 32, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Nagasaka, M.; Shimizu, M.; Tamura, M.; Arata, Y. N-Acetylglucosamine suppresses osteoclastogenesis in part through the promotion of O-GlcNAcylation. Bone Reports 2016, 5, 15–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, T.; Sugimoto, A.; Imazato, N.; Tamura, M.; Nakatani, S.; Kobata, K.; Arata, Y. Glucosamine suppresses osteoclast differentiation through the modulation of glycosylation including O-GlcNAcylation. Biol. Pharm. Bull. 2017, 40, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Wang, L.; Zhu, X.; Lin, W.; Chen, X.; Huang, Z.; Huang, L.; Yang, S. Glucosamine promotes osteoblast proliferation by modulating autophagy via the mammalian target of rapamycin pathway. Biomed. Pharmacother. 2018, 99, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Asai, H.; Nakatani, S.; Kato, T.; Shimizu, T.; Mano, H.; Kobata, K.; Wada, M. Glucosamines attenuate bone loss due to menopause by regulating osteoclast function in ovariectomized mice. Biol. Pharm. Bull. 2016, 39, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Del Mar Arriero, M.; Ramis, J.M.; Perelló, J.; Monjo, M. Inositol hexakisphosphate inhibits osteoclastogenesis on RAW 264.7 Cells and human primary osteoclasts. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Addison, W.N.; McKee, M.D. Inositol hexakisphosphate inhibits mineralization of MC3T3-E1 osteoblast cultures. Bone 2010, 46, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Grases, F.; Sanchis, P.; Prieto, R.M.; Perelló, J.; López-González, Á.A. Effect of tetracalcium dimagnesium phytate on bone characteristics in ovariectomized rats. J. Med. Food 2010, 13, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.C.; Chuang, S.M.; Tang, C.H. D-pinitol inhibits RANKL-induced osteoclastogenesis. Int. Immunopharmacol. 2012, 12, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, S.; Chang, Z.; Kong, D.-X.; Zuo, Z. Quebrachitol: Global Status and basic research. Nat. Products Bioprospect. 2017, 7, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.A.; He, H.; Pham-Huy, C. Chiral drugs: An overview. Int. J. Biomed. Sci. 2006, 2, 85–100. [Google Scholar] [PubMed]
- Wang, Q.; Alén, M.; Nicholson, P.H.F.; Halleen, J.M.; Alatalo, S.L.; Ohlsson, C.; Suominen, H.; Cheng, S. Differential effects of sex hormones on peri- and endocortical bone surfaces in pubertal girls. J. Clin. Endocrinol. MeTable 2006, 91, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.L.; Kang, S.W.; Kang, M.K.; Gong, J.H.; Lee, E.S.; Han, S.J.; Kang, Y.H. Osteoblastogenesis and osteoprotection enhanced by flavonolignan silibinin in osteoblasts and osteoclasts. J. Cell. Biochem. 2012, 113, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.U.; Shin, H.K.; Min, Y.K.; Kim, S.H. Emodin accelerates osteoblast differentiation through phosphatidylinositol 3-kinase activation and bone morphogenetic protein-2 gene expression. Int. Immunopharmacol. 2008, 8, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Oyajobi, B.O.; Harris, S.E.; Chen, D.; Tsao, C.; Deng, H.W.; Zhao, M. Wnt/β-catenin signaling activates bone morphogenetic protein 2 expression in osteoblasts. Bone 2013, 52, 145–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wititsuwannakul, D.; Chareonthiphakorn, N.; Pace, M.; Wititsuwannakul, R. Polyphenol oxidases from latex of Hevea brasiliensis: Purification and characterization. Phytochemistry 2002, 61, 115–121. [Google Scholar] [CrossRef]
- Sritanyarat, W.; Pearce, G.; Siems, W.F.; Ryan, C.A.; Wititsuwannakul, R.; Wititsuwannakul, D. Isolation and characterization of isoinhibitors of the potato protease inhibitor I family from the latex of the rubber trees, Hevea brasiliensis. Phytochemistry 2006, 67, 1644–1650. [Google Scholar] [CrossRef] [PubMed]
- Kanokwiroon, K.; Teanpaisan, R.; Wititsuwannakul, D.; Hooper, A.B.; Wititsuwannakul, R. Antimicrobial activity of a protein purified from the latex of Hevea brasiliensis on oral microorganisms. Mycoses 2008, 51, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Pitakpornpreecha, T.; Plubrukarn, A.; Wititsuwannakul, R. Quantification of 5’-Deoxy-5’-methylthioadenosine in heat-treated natural rubber latex serum. Phytochem. Anal. 2012, 23, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Sukprasirt, P.; Wititsuwannakul, R. A Chitinolytic Endochitinase and β-N-acetylglucosaminidase-based system from Hevea latex in generating N-acetylglucosamine from chitin. Phytochemistry 2014, 104, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Van Alphen, J. Quebrachitol. Ind. Eng. Chem. 1951, 43, 141–145. [Google Scholar] [CrossRef]
- Don, M.J.; Lin, L.C.; Chiou, W.F. Neobavaisoflavone stimulates osteogenesis via p38-mediated up-regulation of transcription factors and osteoid genes expression in MC3T3-E1 Cells. Phytomedicine 2012, 19, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Bronckers, A.L.J.J.; Engelse, M.A.; Cavender, A.; Gaikwad, J.; D’Souza, R.N. Cell-specific patterns of Cbfa1 mRNA and protein expression in postnatal murine dental tissues. Mech. Dev. 2001, 101, 255–258. [Google Scholar] [CrossRef]
- Kim, M.B.; Song, Y.; Hwang, J.K. Kirenol stimulates osteoblast differentiation through activation of the BMP and Wnt/β-catenin signaling pathways in MC3T3-E1 Cells. Fitoterapia 2014, 98, 59–65. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell. 2009, 17, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Monroe, D.G.; McGee-Lawrence, M.E.; Oursler, M.J.; Westendorf, J.J. Update on Wnt signaling in bone cell biology and bone disease. Gene 2012, 492, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, K.I.; Janz, S. Attenuation of WNT signaling by DKK-1 and -2 regulates BMP2-induced osteoblast differentiation and expression of OPG, RANKL and M-CSF. Mol. Cancer 2007, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xu, Y.; Fu, Q.; He, M. Parathyroid hormone regulates osteoblast differentiation in a Wnt/β-catenin-dependent manner. Mol. Cell. Biochem. 2011, 355, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Hipskind, R.A.; Bilbe, G. MAP kinase signaling cascades and gene expression in osteoblasts. Front Biosci. 1998, 3, 804–816. [Google Scholar] [CrossRef]
- Suzuki, A.; Guicheux, J.; Palmer, G.; Miura, Y.; Oiso, Y.; Bonjour, J.P.P.; Caverzasio, J. Evidence for a role of p38 MAP kinase in expression of alkaline phosphatase during osteoblastic cell differentiation. Bone 2002, 30, 91–98. [Google Scholar] [CrossRef]
- Celil, A.B.; Campbell, P.G. BMP-2 and insulin-like growth factor-I mediate osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J. Biol. Chem. 2005, 280, 31353–31359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, C.Y.; Iida-Klein, A.; Wyatt, L.E.; Rudkin, G.H.; Ishida, K.; Yamaguchi, D.T.; Miller, T.A. Serial passage of MC3T3-E1 cells alters osteoblastic function and responsiveness to transforming growth factor-β1 and bone morphogenetic protein-2. Biochem Biophys Res Commun. 1999, 265, 246–251. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |








| Gene | Sequence | GenBank Accession No. |
|---|---|---|
| RUNX2 | F: GTGGCAGTGTCATCATCTGAAAT R: TCGCCTCAGTGATTTAGGGCGCA | NM_001145920.2 |
| OPN | F: GCTATCACCTCGGCCGTTGGGG R: CATTGCCTCCTCCCTCCCGGTG | NM_001204203.1 |
| ALP | F: ATGGAGGATTCCAGATACAGG R: CCATGGTAGATTACGCTCACA | NM_007431.3 |
| OCN | F: CTGTGACATCCATACTTGCAGG R: TGCGCTCTGTCTCTCTGACC | NM_001032298.3 |
| BMP-2 | F: GCTTCCGCTGTTTGTGTTTG R: GGTCACAGATAAGGCCATTGC | NM_007553.3 |
| COLI | F: GCCTTTCCAGGTTCTCCAGCGG R: TTCCCTGGTGCTGATGGTGTTGCT | NM_007742.4 |
| ERK1 | F: AAGCAGAGACCCCAGCAAAGTGAGAGAAG R: GACACCCCTGTCCTTTTGGATCTGGTCCTG | NM_011952.2 |
| ERK2 | F: GTATGGGTGGGCCAGAGCCTGTTCAACTTC R: GGTGCCATGGAACAGGTTGTTCCCAAATGC | NM_001357115.1 |
| JNK1 | F: GCATGGGTCTGATTCTGAAATG R: CTCAGGAGCTCAAGGAATAGTG | NM_001310453.1 |
| JNK2 | F: AACTCTGCGGATGGTGTTC R: GGTCCTCCATAAAGTCCTGTTC | NM_001163671.1 |
| p38α | F: AGGCCATGGTGCATGTGTGT R: AGTAGCTGGAGGAGGAGGAG | NM_001357724.1 |
| LRP5 | F: TTGACCTTGTGGACCCTTTC R: GAGGACAAGCTCCCACATATT | NM_008513.3 |
| β-catenin | F: TCAAGTGAAACCGGGCTATC R: CTCCAACGGGCATCTTCATTA | NM_007664.5 |
| Wnt5a | F: ATATCAGGCACCATTAAACCA R: CACTTAGGGGTTGTTCTCTGA | NM_009524.4 |
| Fzd4 | F: GCACATTGGCACATAAACCGAAC R: GGCTACAACGTGACCAAGATGCC | NM_008055.4 |
| GAPDH | F: GAATTTGCCGTGAGTGGAGT R: AAATGGTGAAGGTCGGTGTG | NM_001289726.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yodthong, T.; Kedjarune-Leggat, U.; Smythe, C.; Wititsuwannakul, R.; Pitakpornpreecha, T. l-Quebrachitol Promotes the Proliferation, Differentiation, and Mineralization of MC3T3-E1 Cells: Involvement of the BMP-2/Runx2/MAPK/Wnt/β-Catenin Signaling Pathway. Molecules 2018, 23, 3086. https://doi.org/10.3390/molecules23123086
Yodthong T, Kedjarune-Leggat U, Smythe C, Wititsuwannakul R, Pitakpornpreecha T. l-Quebrachitol Promotes the Proliferation, Differentiation, and Mineralization of MC3T3-E1 Cells: Involvement of the BMP-2/Runx2/MAPK/Wnt/β-Catenin Signaling Pathway. Molecules. 2018; 23(12):3086. https://doi.org/10.3390/molecules23123086
Chicago/Turabian StyleYodthong, Thanintorn, Ureporn Kedjarune-Leggat, Carl Smythe, Rapepun Wititsuwannakul, and Thanawat Pitakpornpreecha. 2018. "l-Quebrachitol Promotes the Proliferation, Differentiation, and Mineralization of MC3T3-E1 Cells: Involvement of the BMP-2/Runx2/MAPK/Wnt/β-Catenin Signaling Pathway" Molecules 23, no. 12: 3086. https://doi.org/10.3390/molecules23123086