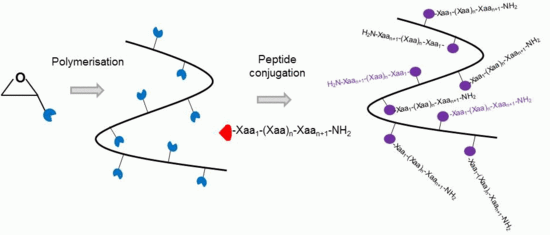
Poly(Ethylene Glycol)-Based Backbones with High Peptide Loading Capacities
Abstract
:1. Introduction
2. Results and Discussion
2.1. Homo- and Co-Polymers of Allyl Glycidyl Ether and Their Functionalisation by the Thiol-Ene Reaction


2.2. PEG Derivatives for Multiple Conjugation of Peptides by Azide-Alkyne Cycloaddition
2.2.1. Alkynylated Polymer and Azido-Peptide




2.2.2. Azido-Polymer and Alkynylated Peptide



2.3. PEG Derivatives for Multiple Conjugation of Peptides by Amide Linkage
Synthesis of Poly(Glycidyl Amine)



2.4. PEG Derivatives for Multiple Conjugations of Peptides by the Thiol-Ene Reaction
2.4.1. Synthesis


2.4.2. Biological Testing

3. Experimental Section
3.1. General Experimental Procedures
3.2. Polymer Synthesis
3.3. General Procedure for Peptide Syntheses
3.4. Conjugations
3.4.1. Cycloaddition Product 7
3.4.2. Cycloaddition Product 11
3.4.3. Amide Linkage Conjugated Product 15
3.4.4. Thiol-Ene Conjugation Product 17
3.5. Biological Assays
3.5.1. Platelet Isolation
3.5.2. Platelets Adhesion Assay
3.5.3. Platelet Aggregation Assay
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef]
- Edwards, C.M.B.; Cohen, M.A.; Bloom, S.R. Peptides as drugs. QJM 1999, 92, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.Y.; Panganiban, B.; Xu, T. Peptide-Polymer conjugates: From fundamental science to application. Annu. Rev. Phys. Chem. 2013, 64, 631–657. [Google Scholar] [CrossRef] [PubMed]
- Chimonides, G.F.; Sohdi, A.A.; Khaleghi, M.R.; Hurley, C.R.; Adams, D.J.; Topham, P.D. Facile synthesis of polymer-peptide conjugates via direct amino acid coupling chemistry. J. Polym. Sci. Polym. Chem. 2013, 51, 4853–4859. [Google Scholar] [CrossRef]
- Veronese, F.M.; Schiavon, O.; Pasut, G.; Mendichi, R.; Andersson, L.; Tsirk, A.; Ford, J.; Wu, G.; Kneller, S.; Davies, J.; et al. PEG-Doxorubicin conjugates: Influence of polymer structure on drug release, in vitro cytotoxicity, biodistribution, and antitumor activity. Bioconjug. Chem. 2005, 16, 775–784. [Google Scholar]
- Hamley, I.W. PEG-Peptide conjugates. Biomacromolecules 2014, 15, 1543–1559. [Google Scholar] [CrossRef] [PubMed]
- Dehn, S.; Chapman, R.; Jolliffe, K.A.; Perrier, S. Synthetic strategies for the design of peptide/polymer conjugates. Polym. Rev. 2011, 51, 214–234. [Google Scholar] [CrossRef]
- Merkel, O.M.; Germershaus, O.; Wada, C.K.; Tarcha, P.J.; Merdan, T.; Kissel, T. Integrin ανβ3 targeted gene delivery using RGD peptidomimetic conjugates with copolymers of PEGylated poly(ethylene imine). Bioconjug. Chem. 2009, 20, 1270–1280. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, S.; Bersani, S.; Scomparin, A.; Mastrotto, F.; Scherpfer, R.; Tonon, G.; Caliceti, P. Tailored PEG for rh-G-CSF analogue site-specific conjugation. Bioconjug. Chem. 2009, 20, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Khalili, H.; Godwin, A.; Choi, J.-W.; Lever, R.; Khaw, P.T.; Brocchini, S. Fab-PEG-Fab as a potential antibody mimetic. Bioconjug. Chem. 2013, 24, 1870–1882. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Hanawa, T.; Nagaoka, S.; Kawakami, H. Design of the complex between manganese porphyrins and catalase-poly(ethylene glycol) conjugates for a new antioxidant. Mol. Pharm. 2007, 4, 484–486. [Google Scholar] [CrossRef] [PubMed]
- Tzokova, N.; Fernyhough, C.M.; Butler, M.F.; Armes, S.P.; Ryan, A.J.; Topham, P.D.; Adams, D.J. The effect of PEO length on the self-assembly of poly(ethylene oxide)—Tetrapeptide conjugates prepared by “click” chemistry. Langmuir 2009, 25, 11082–11089. [Google Scholar] [CrossRef] [PubMed]
- Stutz, C.; Meszynska, A.; Lutz, J.-F.; Börner, H.G. Convenient routes to efficiently N-PEGylated peptides. ACS Macro Lett. 2013, 2, 641–644. [Google Scholar] [CrossRef]
- Mangold, C.; Wurm, F.; Frey, H. Functional PEG-based polymers with reactive groups via anionic ROP of tailor-made epoxides. Polym. Chem. 2012, 3, 1714–1721. [Google Scholar] [CrossRef]
- Lee, B.F.; Kade, M.J.; Chute, J.A.; Gupta, N.; Campos, L.M.; Fredrickson, G.H.; Kramer, E.J.; Lynd, N.A.; Hawker, C.J. Poly(allyl glycidyl ether)—A versatile and functional polyether platform. J. Polym. Sci. Polym. Chem. 2011, 49, 4498–4504. [Google Scholar] [CrossRef]
- Obermeier, B.; Frey, H. Poly(ethylene glycol-co-allyl glycidyl ether)s: A PEG-based modular synthetic platform for multiple bioconjugation. Bioconjug. Chem. 2011, 22, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Mulholland, J.; Nan, A.; McNeill, E.; Ghandehari, H.; Line, B.R. Targeting tumor angiogenic vasculature using polymer—RGD conjugates. J. Control. Release 2005, 102, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, M.A.; Pin, J.-P.R. Interaction of protease-activated receptor 2 with G proteins and β-Arrestin 1 studied by bioluminescence resonance energy transfer. Front. Endocrinol. 2013, 4, 196. [Google Scholar] [CrossRef]
- Cole, M.A.; Jankousky, K.C.; Bowman, C.N. Redox initiation of bulk thiol-ene polymerizations. Polym. Chem. 2013, 4, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Huisgen, R. 1,3-Dipolar cycloadditions. Past and future. Angew. Chem. Int. Ed. 1963, 2, 565–598. [Google Scholar] [CrossRef]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper(I)-Catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Tornoe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1,2,3]-Triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef] [PubMed]
- Thirumurugan, P.; Matosiuk, D.; Jozwiak, K. Click chemistry for drug development and diverse chemical-biology applications. Chem. Rev. 2013, 113, 4905–4979. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, N.C.; Dutta, S.; Datta, M. Mild and efficient deprotection of allyl ethers of phenols and hydroxycoumarins using a palladium on charcoal catalyst and ammonium formate. Tetrahedron Lett. 2006, 47, 5807–5810. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Carpino, L.A.; Han, G.Y. 9-Fluorenylmethoxycarbonyl amino-protecting group. J. Org. Chem. 1972, 37, 3404–3409. [Google Scholar] [CrossRef]
- Hansen, M.B.; van Gurp, T.H.M.; van Hest, J.C.M.; Löwik, D.W.P.M. Simple and efficient solid-phase preparation of azido-peptides. Org. Lett. 2012, 14, 2330–2333. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Neukamm, M.; Metzler-Nolte, N. Synthesis and cytotoxicity of a bimetallic ruthenocene dicobalt-hexacarbonyl alkyne peptide bioconjugate. Dalton Trans. 2011, 40, 1382–1386. [Google Scholar] [CrossRef] [PubMed]
- Cahill, M.; Mistry, R.; Barnett, D.B. The human platelet fibrinogen receptor: Clinical and therapeutic significance. Br. J. Clin. Pharmacol. 1992, 33, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef] [PubMed]
- Hersel, U.; Dahmen, C.; Kessler, H. RGD modified polymers: Biomaterials for stimulated cell adhesion and beyond. Biomaterials 2003, 24, 4385–4415. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.M.; Chess, R.B. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2003, 2, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O'Connor, A.; Marsat, J.-N.; Mitrugno, A.; Flahive, T.; Moran, N.; Brayden, D.; Devocelle, M. Poly(Ethylene Glycol)-Based Backbones with High Peptide Loading Capacities. Molecules 2014, 19, 17559-17577. https://doi.org/10.3390/molecules191117559
O'Connor A, Marsat J-N, Mitrugno A, Flahive T, Moran N, Brayden D, Devocelle M. Poly(Ethylene Glycol)-Based Backbones with High Peptide Loading Capacities. Molecules. 2014; 19(11):17559-17577. https://doi.org/10.3390/molecules191117559
Chicago/Turabian StyleO'Connor, Aoife, Jean-Noel Marsat, Annachiara Mitrugno, Tom Flahive, Niamh Moran, David Brayden, and Marc Devocelle. 2014. "Poly(Ethylene Glycol)-Based Backbones with High Peptide Loading Capacities" Molecules 19, no. 11: 17559-17577. https://doi.org/10.3390/molecules191117559