Structural Characterization of de Novo Designed L5K5W Model Peptide Isomers with Potent Antimicrobial and Varied Hemolytic Activities
Abstract
:Abbreviations
| AMP | antimicrobial peptide |
| CD | circular dichroism |
| CMC | critical micelle concentration |
| COSY | correlated spectroscopy |
| DPC | dodecylphosphocholine |
| GM | geometric mean of the MICs |
| MHC | minimal hemolytic concentration |
| MIC | minimal inhibitory concentration |
| [θ] | mean residue molar ellipticity |
| NATA | N-acetyl-L-tryptophanamide |
| PB | phosphate buffer |
| SDS | sodium dodecylsulfate |
| TFE | trifluoroethanol |
| TI' | pseudo-therapeutic index |
1. Introduction

2. Results and Discussion
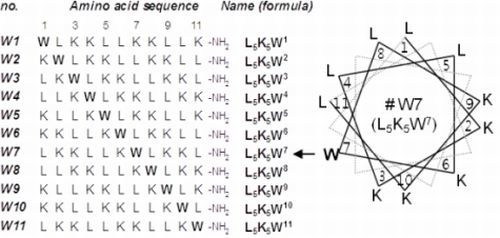
2.1. Peptide Design and Conformational Validation
2.2. Variations in Activities
| W1 a | W2 | W3 | W4 | W5 | W6 | W7 | W8 | W9 | W10 | W11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Minimal inhibitory concentration (μg/mL) | |||||||||||
| Gram-positive bacteria | |||||||||||
| B. subtilis | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 1 | 2 |
| S. aureus | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 |
| S. epidermis | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 4 | 2 | 1 | 2 |
| M. luteus | 4 | 4 | 4 | 4 | 2 | 4 | 4 | 4 | 4 | 2 | 4 |
| Gram-negative bacteria | |||||||||||
| E. coli | 4 | 2 | 4 | 4 | 2 | 2 | 2 | 4 | 2 | 2 | 4 |
| S. dysentariae | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 1 | 2 |
| S. typhimorium | 2 | 2 | 4 | 4 | 2 | 4 | 4 | 4 | 2 | 1 | 4 |
| K. pneumoniae | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 2 | 4 |
| P. aeruginosa | 8 | 8 | 8 | 4 | 8 | 8 | 8 | 8 | 4 | 4 | 4 |
| GM (geometric mean of MICs) | |||||||||||
| Gram (+) b | 2.4 | 2.4 | 2.8 | 2.4 | 2.0 | 2.8 | 2.4 | 3.4 | 2.4 | 1.2 | 2.4 |
| Gram (−) c | 3.0 | 2.3 | 3.5 | 3.5 | 2.6 | 3.5 | 3.0 | 4.6 | 2.3 | 1.7 | 3.5 |
| Gram (+,−) d | 2.7 | 2.3 | 3.2 | 2.9 | 2.3 | 3.2 | 2.7 | 4.0 | 2.3 | 1.5 | 2.9 |
| Methicillin-Resistant S. aureus | |||||||||||
| MRSA-TK784 | 8 | 8 | 8 | 16 | 8 | 4 | 16 | 16 | 8 | 4 | 16 |

| Peptides | MHC (μg/mL) | TI' | |||
|---|---|---|---|---|---|
| (+) a | (−) b | (+, −) c | |||
| L5K5W1 (W1) | 32 | 13.5 | 10.6 | 11.8 | |
| L5K5W2 (W2) | 16 | 6.7 | 7.0 | 6.9 | |
| L5K5W3 (W3) | 64 | 22.6 | 18.4 | 20.2 | |
| L5K5W4 (W4) | 128 | 53.8 | 36.8 | 43.5 | |
| L5K5W5 (W5) | 64 | 32.0 | 24.3 | 27.4 | |
| L5K5W6 (W6) | 32 | 11.3 | 9.2 | 10.1 | |
| L5K5W7 (W7) | 128 | 53.8 | 42.2 | 47.0 | |
| L5K5W8 (W8) | 128 | 38.1 | 27.9 | 32.0 | |
| L5K5W9 (W9) | 64 | 26.9 | 27.9 | 27.4 | |
| L5K5W10 (W10) | 32 | 26.9 | 18.4 | 21.8 | |
| L5K5W11 (W11) | 128 | 53.8 | 36.8 | 43.5 | |
2.3. Conformational Behavior

2.4. Tryptophanyl Environments


2.5. Structure-Activity Relationships

3. Experimental
3.1. Peptide Preparation
3.2. Circular Dichroism (CD) Spectroscopy
3.3. Nuclear Magnetic Resonance (NMR) Spectroscopy
3.4. Fluorescence Spectroscopy
3.5. Antimicrobial Assay
3.6. Hemolytic Assay
4. Conclusions
Acknowledgments
- Sample Availability: Samples of the W1-W11 peptides are available from the authors.
References
- Nakatsuji, T.; Gallo, R.L. Antimicrobial peptides: Old molecules with new ideas. J. Invest. Dermatol. 2012, 132, 887–895. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Wiesner, J.; Vilcinskas, A. Antimicrobial peptides: The ancient arm of the human immune system. Virulence 2010, 1, 440–464. [Google Scholar] [CrossRef]
- Zaiou, M. Multifunctional antimicrobial peptides: Therapeutic targets in several human diseases. J. Mol. Med. 2007, 85, 317–329. [Google Scholar] [CrossRef]
- Bevins, C.L.; Salzman, N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbial. 2011, 9, 356–368. [Google Scholar] [CrossRef]
- Frew, L.; Stock, S.J. Antimicrobial peptides and pregnancy. Reproduction 2011, 141, 725–735. [Google Scholar] [CrossRef]
- Schröder, J.M. Antimicrobial peptides in healthy skin and atopic dermatitis. Allergol. Int. 2011, 60, 17–24. [Google Scholar] [CrossRef]
- Gorr, S.U. Antimicrobial peptides in periodontal innate defense. Front Oral Biol. 2012, 15, 84–98. [Google Scholar] [CrossRef]
- Su, Y.; Zhang, K.; Schluesener, H.J. Antimicrobial peptides in the brain. Arch. Immunol. Ther. Exp. 2010, 58, 365–377. [Google Scholar] [CrossRef]
- Marta-Guarna, M.; Coulson, R.; Rubinchik, E. Anti-inflammatory activity of cationic peptide: Application to the treatment of acne vulgaris. FEMS Microbiol. Lett. 2006, 257, 1–6. [Google Scholar] [CrossRef]
- Scott, M.G.; Hancock, R.E. Cationic antimicrobial peptides and their multifunctional role in the immune system. Crit. Rev. Immunol. 2000, 20, 407–431. [Google Scholar]
- Mader, J.S.; Hoskin, D.W. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert Opin. Investig. Drugs 2006, 15, 933–946. [Google Scholar] [CrossRef]
- Papo, N.; Shai, Y. Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. 2005, 62, 784–790. [Google Scholar] [CrossRef]
- Gubern, C.; Lόpez-Bermejo, A.; Biarnés, J.; Vendrell, J.; Ricart, W.; Fernández-Real, J.M. Natural antibiotics and insulin sensitivity: The role of bactericidal/permeability-increasing protein. Diabetes 2006, 55, 216–224. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, J.O.; Jung, J.H.; Lee, S.K.; You, G.Y.; Park, S.J.; Kim, H.S. Gaegurin-6 stimulates insulin secretion through calcium influx in pancreatic β Rin5mf cells. Regul. Pept. 2009, 159, 123–128. [Google Scholar]
- Rotem, S.; Mor, A. Antimicrobial peptide mimics for improved therapeutic properties. Biochim. Biophys. Acta 2009, 1788, 1582–1592. [Google Scholar] [CrossRef]
- Brogden, N.K.; Brogden, K.A. Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int. J. Antimicrob. Agents 2011, 38, 217–225. [Google Scholar]
- Baltzer, S.A.; Brown, M.H. Antimicrobial peptides—Promising alternatives to conventional antibiotics. J. Mol. Microbiol. Biotechnol. 2011, 20, 228–235. [Google Scholar] [CrossRef]
- Oyston, P.C.F.; Fox, M.A.; Richards, S.J.; Clark, G.C. Novel peptides therapeutics for treatment of infections. J. Med. Microbiol. 2009, 58, 977–987. [Google Scholar] [CrossRef]
- Lee, S.-H.; Kim, S.-J.; Lee, Y.-S.; Song, M.-D.; Kim, I.-H.; Won, H.-S. De novo generation of short antimicrobial peptides with simple amino acid composition. Regul. Pept. 2011, 166, 36–41. [Google Scholar] [CrossRef]
- Kang, S.-J.; Won, H.-S.; Choi, W.-S.; Lee, B.-J. De novo generation of antimicrobial LK peptides with a single tryptophan at the critical amphipathic interface. J. Pept. Sci. 2009, 15, 583–588. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef]
- Zelezetsky, I.; Tossi, A. Alpha-helical antimicrobial peptides—Using a sequence template to guide structure-activity relationships studies. Biochim. Biophys. Acta 2006, 1758, 1436–1449. [Google Scholar] [CrossRef]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic α helical antimicrobial peptides: A systematic study of the effects of structural and physical properties on biological activity. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, α-helical antimicrobial peptides. Biopolymers 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Won, H.-S.; Kang, S.-J.; Lee, B.-J. Action mechanism and structural requirements of the antimicrobial peptides, gaegurins. Biochim. Biophys. Acta 2009, 1788, 1620–1629. [Google Scholar] [CrossRef]
- Won, H.-S.; Park, S.-H.; Kim, H.E.; Hyun, B.; Kim, M.; Lee, B.J.; Lee, B.-J. Effects of a tryptophanyl substitution on the structure and antimicrobial activity of C-terminally truncated gaegurin 4. Eur. J. Biochem. 2002, 269, 4367–4374. [Google Scholar] [CrossRef]
- Won, H.-S.; Jung, S.-J.; Kim, H.-E.; Seo, M.-D.; Lee, B.-J. Systematic peptide engineering and structural characterization to search for the shortest antimicrobial peptide analogue of gaegurin 5. J. Biol. Chem. 2004, 279, 14784–14791. [Google Scholar]
- Won, H.-S.; Seo, M.-D.; Jung, S.-J.; Lee, S.-J.; Kang, S.-J.; Son, W.-S.; Kim, H.-J.; Park, T.-K.; Park, S.-J.; Lee, B.-J. Structural determinants for the membrane interaction of novel bioactive undecapeptides derived from gaegurin 5. J. Med. Chem. 2006, 49, 4886–4895. [Google Scholar] [CrossRef]
- Won, H.-S.; Kang, S.-J.; Choi, W.-S.; Lee, B.-J. Activity optimization of an undecapeptide analogue derived from a frog-skin antimicrobial peptide. Mol. Cells 2011, 31, 49–54. [Google Scholar] [CrossRef]
- Chou, P.Y.; Fasman, G.D. Conformational parameters for amino acids in helical, β-sheet, and random coil regions calculated from proteins. Biochemistry 1974, 13, 211–222. [Google Scholar]
- Pace, C.N.; Scholtz, J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998, 75, 422–427. [Google Scholar] [CrossRef]
- Schiffer, M.; Chang, C.H.; Stevens, F.J. The functions of tryptophan residues in membrane proteins. Protein Eng. 1992, 5, 213–214. [Google Scholar] [CrossRef]
- Ridder, A.N.; Morein, S.; Stam, J.G.; Kuhn, A.; de Kruijff, B.; Killian, J.A. Analysis of the role of interfacial tryptophan residues in controlling the topology of membrane proteins. Biochemistry 2000, 39, 6521–6528. [Google Scholar] [CrossRef]
- Zhang, X.; Adda, C.G.; Low, A.; Zhang, J.; Zhang, W.; Sun, H.; Tu, X.; Anders, R.F.; Norton, R.S. Role of the helical structure of the N-terminal region of Plasmodium falciparummerozoite surface protein 2 in fibril formation and membrane interaction. Biochemistry 2012, 51, 1380–1387. [Google Scholar]
- Tulumello, D.V.; Deber, C.M. SDS micelles as a membrane-mimetic environment for transmembrane segments. Biochemistry 2009, 48, 12096–12103. [Google Scholar] [CrossRef]
- Sanders, C.R.; Sönnichsen, F. Solution NMR of membrane proteins: Practice and challenges. Magn. Reson. Chem. 2006, 44, 24–40. [Google Scholar] [CrossRef]
- Javadpour, M.M.; Barkley, M.D. Self-assembly of designed antimicrobial peptides in solution and micelles. Biochemistry 1997, 36, 9540–9549. [Google Scholar] [CrossRef]
- Zhu, W.L.; Nan, Y.H.; Hahm, K.S.; Shin, S.Y. Cell selectivity of an antimicrobial peptide melittindiastereomer with D-amino acid in the leucine zipper sequence. J. Biochem. Mol. Biol. 2007, 40, 1090–1094. [Google Scholar] [CrossRef]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Rational design of α-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar]
- Ubukata, K.; Yamashita, N.; Konno, M. Occurrence of a β-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 1985, 5, 851–857. [Google Scholar]
- Badosa, E.; Ferre, R.; Planas, M.; Feliu, L.; Besalú, E.; Cabrefiga, J.; Bardají, E.; Montesinos, E. A library of linear undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides 2007, 28, 2276–2285. [Google Scholar] [CrossRef]
- Ferre, R.; Badosa, E.; Feliu, L.; Planas, M.; Montesinos, E.; Bardají, E. Inhibition of plant-pathogenic bacteria by short synthetic cecropin A-melittin hybrid peptides. Appl. Environ. Microbiol. 2006, 72, 3302–3308. [Google Scholar]
- Holtzer, M.E.; Holtzer, A. Alpha-helix to random coil transitions: Determination of peptide concentration from the CD at the isodichroic point. Biopolymers 1992, 32, 1675–1677. [Google Scholar] [CrossRef]
- Storrs, R.W.; Truckses, D.; Wemmer, D.E. Helix propagation in trifluoroethanol solutions. Biopolymers 1992, 32, 1695–1702. [Google Scholar] [CrossRef]
- Khandelwal, P.; Seth, S.; Hosur, R.V. CD and NMR investigations on trifluoroethanol-induced step-wise folding of helical segment from scorpion neurotoxin. Eur. J. Biochem. 1999, 264, 468–478. [Google Scholar] [CrossRef]
- Buchko, G.W.; Rozek, A.; Hoyt, D.W.; Cushley, R.J.; Kennedy, M.A. The use of sodium dodecyl sulfate to model the apolipoprotein environment. Evidence for peptide-SDS complexes using pulsed-field-gradient NMR spectroscopy. Biochim. Biophys. Acta 1998, 1392, 101–108. [Google Scholar] [CrossRef]
- Arnold, G.E.; Day, L.A.; Dunker, A.K. Tryptophan contributions to the unusual circular dichroism of fdbacteriophage. Biochemistry 1992, 31, 7948–7956. [Google Scholar] [CrossRef]
- Franke, J.D.; Dong, F.; Rickoll, W.L.; Kelley, M.J.; Kiehart, D.P. Rod mutations associated with MYH9-related disorders disrupt nonmuscle myosin-IIA assembly. Blood 2005, 105, 161–169. [Google Scholar] [CrossRef]
- Stafford, R.E.; Fanni, T.; Dennis, E.A. Interfacial properties and critical micelle concentration of lysophospholipids. Biochemistry 1989, 28, 5113–5120. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar]
- Dathe, M.; Meyer, J.; Beyermann, M.; Maul, B.; Hoischen, C.; Bienert, M. General aspects of peptide selectivity towards lipid bilayers and cell membranes studied by variation of the structural parameters of amphipathic helical model peptides. Biochim. Biophys. Acta 2002, 1558, 171–186. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Kim, S.; Kim, S.-S.; Lee, B.-J. Correlation between the activities of alpha-helical antimicrobial peptides and hydrophobicities represented as RP HPLC retention times. Peptides 2005, 26, 2050–2056. [Google Scholar] [CrossRef]
- Eisenberg, D. Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. 1984, 53, 595–623. [Google Scholar] [CrossRef]
- Greenfield, N.J. Applications of circular dichroism in protein and peptide analysis. Trends Analyt. Chem. 1999, 18, 236–244. [Google Scholar] [CrossRef]
- Böhm, G.; Muhr, R.; Jaenicke, R. Quantitative analysis of protein far UV circular dichroism spectra by neural network. Protein Eng. 1992, 5, 191–195. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kim, S.-J.; Kim, J.-S.; Lee, Y.-S.; Sim, D.-W.; Lee, S.-H.; Bahk, Y.-Y.; Lee, K.-H.; Kim, E.-H.; Park, S.-J.; Lee, B.-J.; et al. Structural Characterization of de Novo Designed L5K5W Model Peptide Isomers with Potent Antimicrobial and Varied Hemolytic Activities. Molecules 2013, 18, 859-876. https://doi.org/10.3390/molecules18010859
Kim S-J, Kim J-S, Lee Y-S, Sim D-W, Lee S-H, Bahk Y-Y, Lee K-H, Kim E-H, Park S-J, Lee B-J, et al. Structural Characterization of de Novo Designed L5K5W Model Peptide Isomers with Potent Antimicrobial and Varied Hemolytic Activities. Molecules. 2013; 18(1):859-876. https://doi.org/10.3390/molecules18010859
Chicago/Turabian StyleKim, Seo-Jin, Jae-Seok Kim, Yoo-Sup Lee, Dae-Won Sim, Sung-Hee Lee, Young-Yil Bahk, Kwang-Ho Lee, Eun-Hee Kim, Sung-Jean Park, Bong-Jin Lee, and et al. 2013. "Structural Characterization of de Novo Designed L5K5W Model Peptide Isomers with Potent Antimicrobial and Varied Hemolytic Activities" Molecules 18, no. 1: 859-876. https://doi.org/10.3390/molecules18010859