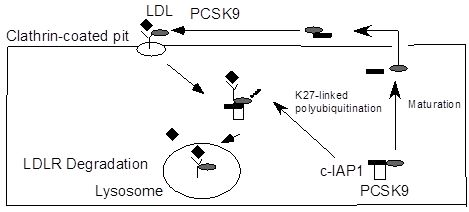
c-IAP1 Binds and Processes PCSK9 Protein: Linking the c-IAP1 in a TNF-α Pathway to PCSK9-Mediated LDLR Degradation Pathway
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Potential PCSK9 Binding Proteins through Affinity Purification and Shotgun LC-MS/MS Analysis
| Gene Symbols | Gene Names | Experiment 1 | Experiment 2 | Experiment 3 | |||
|---|---|---|---|---|---|---|---|
| emp | exp | emp | exp | emp | exp | ||
| PCSK9 | proprotein convertase subtilisin/kexin type 9 | 3 | 1466 | 2 | 1171 | 4 | 1128 |
| UGGT1 | UDP-glucose glycoprotein glucosyltransferase 1 | 0 | 9 | 0 | 12 | 0 | 15 |
| PDIA4 (ERP72) | protein disulfide isomerase family A, member 4 | 0 | 7 | 0 | 5 | 0 | 7 |
| SLC25A1 | solute carrier family 25 member 1 | 0 | 6 | 0 | 2 | 0 | 6 |
| DNAJA1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | 0 | 12 | 0 | 10 | 0 | 7 |
| CLGN | Calmegin | 0 | 2 | 0 | 2 | 0 | 2 |
| DNAJA2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 0 | 2 | 0 | 3 | 0 | 4 |
| c-IAP1 (BIRC2) | cellular inhibitor of apoptosis protein 1 | 0 | 7 | 0 | 9 | 0 | 8 |
| TBB6 | tubulin, beta 6 | 0 | 5 | 0 | 2 | 0 | 3 |
| TRAF2 | TNF receptor-associated factor 2 | 0 | 4 | 0 | 3 | 0 | 3 |
| SLC25A10 | solute carrier family 25. member 10 | 0 | 6 | 0 | 3 | 0 | 6 |
| DNAJB11 | DnaJ (Hsp40) homolog, subfamily B, member 11 | 0 | 5 | 0 | 4 | 0 | 3 |
| CDIPT | CDP-diacylglycerol-inositol 3-phosphatidyltransferase | 0 | 3 | 0 | 6 | 0 | 4 |
| DNAJC10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | 0 | 6 | 0 | 3 | 0 | 6 |
| DNAJA3 | DnaJ (Hsp40) homolog, subfamily A, member 3 | 0 | 2 | 0 | 3 | 0 | 3 |
| RHOT1 | ras homolog gene family, member T1 | 0 | 2 | 0 | 3 | 0 | 4 |
| AGK | Acylglycerol kinase lipid kinase | 0 | 2 | 0 | 3 | 0 | 2 |
| HSPB1 | heat shock 27kDa protein 1 | 0 | 4 | 0 | 5 | 0 | 2 |
| RCN1 | reticulocalbin 1 | 0 | 3 | 0 | 4 | 0 | 5 |
| Ribosomal L28 | partial ribosomal protein L28 variant | 0 | 2 | 0 | 6 | 0 | 5 |
| Stub1 | STIP1 homology and U-box containing protein 1 | 0 | 2 | 0 | 3 | 0 | 2 |
| PGRC1 | progesterone receptor membrane component 1 | 0 | 2 | 0 | 2 | 0 | 2 |
| SLC25A12 | Solute carrier family 25 member 12 | 0 | 2 | 0 | 5 | 0 | 4 |
2.2. c-IAP1/TRAF2 Complex Binds PCSK9


2.3. c-IAP1 Knock-Down
2.4. Ubiquitination of PCSK9 by c-IAP1


2.5. Discussion
3. Experimental
3.1. Cell Culture and Immunofluorescence Staining and Image Analysis
3.2. DNA Constructs, Transfections and Western Blot Assays
3.3. Generation of Stable Cell Lines, Immunoprecipitation (IP) and Protein Purification
3.4. Shotgun Analysis of the FLAG-Tagged PCSK9 and Associated Proteins Complex Samples
3.5. RNA Interference
3.6. Mouse PCSK9 Immunoassay
3.7. In Vivo Ubiquitination Analysis
3.8. In Vitro Ubiquitination Assay
3.9. Mass Spectrometric Analysis of Ploy-Ubiquitinated PCSK9
4. Conclusions
Supplementary Materials
Acknowledgements
Authors’ Contributions
- Sample Availability: Samples are available from the corresponding author, Dr. Weiming Xu, Email: [email protected].
References
- Abifadel, M.; Varret, M.; Rabes, J.P.; Allard, D.; Ouguerram, K.; Devillers, M.; Cruaud, C.; Benjannet, S.; Wickham, L.; Erlich, D.; et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003, 34, 154–156. [Google Scholar] [CrossRef]
- Maxwell, K.N.; Breslow, J.L. Proprotein convertase subtilisin kexin 9: The third locus implicated in autosomal dominant hypercholesterolemia. Curr. Opin. Lipidol. 2005, 16, 167–172. [Google Scholar] [CrossRef]
- Cohen, J.C.; Boerwinkle, E.; Mosley, T.H., Jr.; Hobbs, H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006, 354, 1264–1272. [Google Scholar] [CrossRef]
- Costet, P.; Krempf, M.; Cariou, B. PCSK9 and LDL cholesterol: Unravelling the target to design the bullet. Trends Biochem. Sci. 2008, 33, 426–434. [Google Scholar] [CrossRef]
- Horton, J.D.; Cohen, J.C.; Hobbs, H.H. Molecular biology of PCSK9: Its role in LDL metabolism. Trends Biochem. Sci. 2007, 32, 71–77. [Google Scholar] [CrossRef]
- Seidah, N.G.; Prat, A. The proprotein convertases are potential targets in the treatment of dyslipidemia. J. Mol. Med. 2007, 85, 685–696. [Google Scholar] [CrossRef]
- Poirier, S.; Mayer, G.; Benjannet, S.; Bergeron, E.; Marcinkiewicz, J.; Nassoury, N.; Mayer, H.; Nimpf, J.; Prat, A.; Seidah, N.G. The proprotein convertase PCSK9 induces the degradation of low density lipoprotein receptor (LDLR) and its closest family members VLDLR and ApoER2. J. Biol. Chem. 2008, 283, 2363–2372. [Google Scholar]
- Park, S.W.; Moon, Y.A.; Horton, J.D. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J. Biol. Chem. 2004, 279, 50630–50638. [Google Scholar]
- Liu, H.; Sadygov, R.G.; Yates, J.R., 3rd. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004, 76, 4193–4201. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Ashwell, J.D. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 2002, 416, 345–347. [Google Scholar] [CrossRef]
- Vince, J.E.; Wong, W.W.; Khan, N.; Feltham, R.; Chau, D.; Ahmed, A.U.; Benetatos, C.A.; Chunduru, S.K.; Condon, S.M.; McKinlay, M.; et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 2007, 131, 682–693. [Google Scholar] [CrossRef]
- Kamitani, T.; Kito, K.; Nguyen, H.P.; Yeh, E.T. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 1997, 272, 28557–28562. [Google Scholar] [CrossRef]
- Denis, N.J.; Vasilescu, J.; Lambert, J.P.; Smith, J.C.; Figeys, D. Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics 2007, 7, 868–874. [Google Scholar] [CrossRef]
- Lagace, T.A.; Curtis, D.E.; Garuti, R.; McNutt, M.C.; Park, S.W.; Prather, H.B.; Anderson, N.N.; Ho, Y.K.; Hammer, R.E.; Horton, J.D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 2006, 116, 2995–3005. [Google Scholar] [CrossRef]
- Ikeda, H.; Kerppola, T.K. Lysosomal localization of ubiquitinated Jun requires multiple determinants in a lysine-27-linked polyubiquitin conjugate. Mol. Biol. Cell 2008, 19, 4588–4601. [Google Scholar] [CrossRef]
- Wu, C.J.; Conze, D.B.; Li, X.; Ying, S.X.; Hanover, J.A.; Ashwell, J.D. TNF-alpha induced c-IAP1/TRAF2 complex translocation to a Ubc6-containing compartment and TRAF2 ubiquitination. EMBO J. 2005, 24, 1886–1898. [Google Scholar] [CrossRef]
- Vince, J.E.; Chau, D.; Callus, B.; Wong, W.W.; Hawkins, C.J.; Schneider, P.; McKinlay, M.; Benetatos, C.A.; Condon, S.M.; Chunduru, S.K.; et al. TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1-TRAF2 complex to sensitize tumor cells to TNFalpha. J. Cell Biol. 2008, 182, 171–184. [Google Scholar] [CrossRef]
- Afshar, N.; Black, B.E.; Paschal, B.M. Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell Biol. 2005, 25, 8844–8853. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Berge, K.E.; Leren, T.P. The unique role of proprotein convertase subtilisin/kexin 9 in cholesterol homeostasis. J. Intern. Med. 2009, 266, 507–519. [Google Scholar] [CrossRef]
- Choi, Y.E.; Butterworth, M.; Malladi, S.; Duckett, C.S.; Cohen, G.M.; Bratton, S.B. The E3 ubiquitin ligase cIAP1 binds and ubiquitinates caspase-3 and -7 via unique mechanisms at distinct steps in their processing. J. Biol. Chem. 2009, 284, 12772–12782. [Google Scholar]
- Ghribi, O.; Herman, M.M.; Savory, J. The endoplasmic reticulum is the main site for caspase-3 activation following aluminum-induced neurotoxicity in rabbit hippocampus. Neurosci. Lett. 2002, 324, 217–221. [Google Scholar] [CrossRef]
- McNutt, M.C.; Lagace, T.A.; Horton, J.D. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J. Biol. Chem. 2007, 282, 20799–20803. [Google Scholar]
- Pandit, S.; Wisniewski, D.; Santoro, J.C.; Ha, S.; Ramakrishnan, V.; Cubbon, R.M.; Cummings, R.T.; Wright, S.D.; Sparrow, C.P.; Sitlani, A.; et al. Functional analysis of sites within PCSK9 responsible for hypercholesterolemia. J. Lipid Res. 2008, 49, 1333–1343. [Google Scholar] [CrossRef]
- Kwon, H.J.; Lagace, T.A.; McNutt, M.C.; Horton, J.D.; Deisenhofer, J. Molecular basis for LDL receptor recognition by PCSK9. Proc. Natl. Acad. Sci. USA 2008, 105, 1820–1825. [Google Scholar]
- Ballinger, C.A.; Connell, P.; Wu, Y.; Hu, Z.; Thompson, L.J.; Yin, L.Y.; Patterson, C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell Biol. 1999, 19, 4535–4545. [Google Scholar]
- Welchman, R.L.; Gordon, C.; Mayer, R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005, 6, 599–609. [Google Scholar] [CrossRef]
- Zelcer, N.; Hong, C.; Boyadjian, R.; Tontonoz, P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 2009, 325, 100–104. [Google Scholar]
- Xu, W.; Liu, L.; Charles, I.G.; Moncada, S. Nitric oxide induces coupling of mitochondrial signalling with the endoplasmic reticulum stress response. Nat. Cell Biol. 2004, 6, 1129–1134. [Google Scholar] [CrossRef]
- Xu, W.; Liu, L.; Brown, N.J.; Christian, S.; Hornby, D. Quantum dot-conjugated anti-GRP78 scFv inhibits cancer growth in mice. Molecules 2012, 17, 796–808. [Google Scholar] [CrossRef]
- Livingston, C.M.; Ifrim, M.F.; Cowan, A.E.; Weller, S.K. Virus-Induced Chaperone-Enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 2009, 5, e1000619. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xu, W.; Liu, L.; Hornby, D. c-IAP1 Binds and Processes PCSK9 Protein: Linking the c-IAP1 in a TNF-α Pathway to PCSK9-Mediated LDLR Degradation Pathway. Molecules 2012, 17, 12086-12101. https://doi.org/10.3390/molecules171012086
Xu W, Liu L, Hornby D. c-IAP1 Binds and Processes PCSK9 Protein: Linking the c-IAP1 in a TNF-α Pathway to PCSK9-Mediated LDLR Degradation Pathway. Molecules. 2012; 17(10):12086-12101. https://doi.org/10.3390/molecules171012086
Chicago/Turabian StyleXu, Weiming, Lizhi Liu, and David Hornby. 2012. "c-IAP1 Binds and Processes PCSK9 Protein: Linking the c-IAP1 in a TNF-α Pathway to PCSK9-Mediated LDLR Degradation Pathway" Molecules 17, no. 10: 12086-12101. https://doi.org/10.3390/molecules171012086