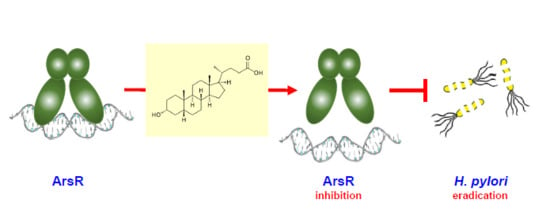
Small Molecule Inhibitors of the Response Regulator ArsR Exhibit Bactericidal Activity against Helicobacter pylori
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Media and Growth Conditions
2.2. Chemicals
2.3. Recombinant Expression and Purification of the H. pylori Response Regulator ArsR
2.4. High-Throughput Screening for ArsR Binders
2.5. Electrophoretic Mobility Shift Assays
2.6. Isothermal Titration Calorimetry Assays
2.7. Minimal Inhibitory and Bactericidal Concentrations
2.8. Time-Kill Kinetics Assays
2.9. Checkerboard Assays
3. Results
3.1. Screening of a Repurposing Chemical Library Identified Several ArsR Binders
3.2. Some Non-Antibiotic FDA-Approved Drugs Inhibited the DNA Binding Activity of ArsR
3.3. ArsR Inhibitors Bound to the Protein at 1:1 Stoichiometry in the Micromolar Range
3.4. ArsR Inhibitors Exhibited Bactericidal Activities against H. pylori
3.5. Lithocholic Acid Exhibits Low Antimicrobial Activities against Members of Human Normal Microbiota
3.6. Lithocholic Acid Partially Synergizes with Other Antimicrobial Drugs against H. pylori
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Medina, E.; Pieper, D.H. Tackling threats and future problems of multidrug-resistant bacteria. Curr. Top. Microbiol. Immunol. 2016, 398, 3–33. [Google Scholar] [CrossRef] [PubMed]
- Fields, F.R.; Lee, S.W.; McConnell, M.J. Using bacterial genomes and essential genes for the development of new antibiotics. Biochem. Pharmacol. 2017, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Fillat, M.F.; Lanas, A. Transcriptional regulators: Valuable targets for novel antibacterial strategies. Future Med. Chem. 2018, 10, 541–560. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Pflock, M.; Finsterer, N.; Joseph, B.; Mollenkopf, H.; Meyer, T.F.; Beier, D. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 2006, 188, 3449–3462. [Google Scholar] [CrossRef] [Green Version]
- Muller, S.; Gotz, M.; Beier, D. Histidine residue 94 is involved in pH sensing by histidine kinase ArsS of Helicobacter pylori. PLoS ONE 2009, 4, e6930. [Google Scholar] [CrossRef] [Green Version]
- Marcus, E.A.; Sachs, G.; Wen, Y.; Scott, D.R. Phosphorylation-dependent and phosphorylation-independent regulation of Helicobacter pylori acid acclimation by the ArsRS two-component system. Helicobacter 2016, 21, 69–81. [Google Scholar] [CrossRef]
- Pflock, M.; Kennard, S.; Delany, I.; Scarlato, V.; Beier, D. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 2005, 73, 6437–6445. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.; Feng, J.; Scott, D.R.; Marcus, E.A.; Sachs, G. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 alpha-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J. Bacteriol. 2007, 189, 2426–2434. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, A.C.; Weinberger, D.M.; Ford, C.B.; Nelson, J.C.; Snider, J.D.; Hall, J.D.; Paules, C.I.; Peek, R.M., Jr.; Forsyth, M.H. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology 2008, 154, 2231–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servetas, S.L.; Carpenter, B.M.; Haley, K.P.; Gilbreath, J.J.; Gaddy, J.A.; Merrell, D.S. Characterization of key Helicobacter pylori regulators identifies a role for ArsRS in biofilm formation. J. Bacteriol. 2016, 198, 2536–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servetas, S.L.; Doster, R.S.; Kim, A.; Windham, I.H.; Cha, J.H.; Gaddy, J.A.; Merrell, D.S. ArsRS-dependent regulation of homB contributes to Helicobacter pylori biofilm formation. Front. Microbiol. 2018, 9, 1497. [Google Scholar] [CrossRef] [PubMed]
- Acio-Pizzarello, C.R.; Acio, A.A.; Choi, E.J.; Bond, K.; Kim, J.; Kenan, A.C.; Chen, J.; Forsyth, M.H. Determinants of the regulation of Helicobacter pylori adhesins include repeat sequences in both promoter and coding regions as well as the two-component system ArsRS. J. Med. Microbiol. 2017, 66, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Beier, D.; Frank, R. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 2000, 182, 2068–2076. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Salillas, S.; Velázquez-Campoy, A.; Espinosa Angarica, V.; Fillat, M.F.; Sancho, J.; Lanas, Á. Identifying potential novel drugs against Helicobacter pylori by targeting the essential response regulator HsrA. Sci. Rep. 2019, 9, 11294. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Casado, J.; Chueca, E.; Salillas, S.; Velázquez-Campoy, A.; Espinosa Angarica, V.; Benejat, L.; Guignard, J.; Giese, A.; Sancho, J.; et al. Repurposing dihydropyridines for treatment of Helicobacter pylori infection. Pharmaceutics 2019, 11, 681. [Google Scholar] [CrossRef] [Green Version]
- Cremades, N.; Velázquez-Campoy, A.; Martínez-Júlvez, M.; Neira, J.L.; Pérez-Dorado, I.; Hermoso, J.; Jiménez, P.; Lanas, A.; Hoffman, P.S.; Sancho, J. Discovery of specific flavodoxin inhibitors as potential therapeutic agents against Helicobacter pylori infection. ACS Chem. Biol. 2009, 4, 928–938. [Google Scholar] [CrossRef]
- Velázquez-Campoy, A.; Sancho, J.; Abián, O.; Vega, S. Biophysical screening for identifying pharmacological chaperones and inhibitors against conformational and infectious diseases. Curr. Drug Targets 2016, 17, 1492–1505. [Google Scholar] [CrossRef] [Green Version]
- Sancho, J. The stability of 2-state, 3-state and more-state proteins from simple spectroscopic techniques... plus the structure of the equilibrium intermediates at the same time. Arch. Biochem. Biophys. 2013, 531, 4–13. [Google Scholar] [CrossRef]
- Dietz, P.; Gerlach, G.; Beier, D. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 2002, 184, 350–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velázquez-Campoy, A.; Leavitt, S.A.; Freire, E. Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol. Biol. 2015, 1278, 183–204. [Google Scholar] [CrossRef] [PubMed]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society for Clinical Microbiology and Infectious Diseases (ESCMID). EUCAST Discussion Document E. Dis 5.1: Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, 1–7. [Google Scholar]
- Krzyzek, P.; Franiczek, R.; Krzyzanowska, B.; Laczmanski, L.; Migdal, P.; Gosciniak, G. In vitro activity of 3-Bromopyruvate, an anticancer compound, against antibiotic-susceptible and antibiotic-resistant Helicobacter pylori strains. Cancers 2019, 11, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimmperman, P.; Baranauskiene, L.; Jachimoviciute, S.; Jachno, J.; Torresan, J.; Michailoviene, V.; Matuliene, J.; Sereikaite, J.; Bumelis, V.; Matulis, D. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys. J. 2008, 95, 3222–3231. [Google Scholar] [CrossRef] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: http://www.eucast.org (accessed on 30 March 2020).
- Banerjee, A.; Majumder, P.; Sanyal, S.; Singh, J.; Jana, K.; Das, C.; Dasgupta, D. The DNA intercalators ethidium bromide and propidium iodide also bind to core histones. FEBS Open Bio 2014, 4, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Cheesman, M.J.; Ilanko, A.; Blonk, B.; Cock, I.E. Developing new antimicrobial therapies: Are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn. Rev. 2017, 11, 57–72. [Google Scholar] [CrossRef] [Green Version]
- Falagas, M.E.; Bliziotis, I.A. Pandrug-resistant Gram-negative bacteria: The dawn of the post-antibiotic era? Int. J. Antimicrob. Agents 2007, 29, 630–636. [Google Scholar] [CrossRef]
- Gill, J.S.; Arora, S.; Khanna, S.P.; Kumar, K.H. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level intensive care unit. J. Glob. Infect. Dis. 2016, 8, 155–159. [Google Scholar]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundback, T.; Nordlund, P.; Martinez Molina, D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef]
- Galano, J.J.; Alias, M.; Perez, R.; Velazquez-Campoy, A.; Hoffman, P.S.; Sancho, J. Improved flavodoxin inhibitors with potential therapeutic effects against Helicobacter pylori infection. J. Med. Chem. 2013, 56, 6248–6258. [Google Scholar] [CrossRef] [PubMed]
- Berg, T. Inhibition of transcription factors with small organic molecules. Curr. Opin. Chem. Biol. 2008, 12, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, Y.; Doi, A.; Furuta, E.; Dubrac, S.; Ishizaki, Y.; Okada, M.; Igarashi, M.; Misawa, N.; Yoshikawa, H.; Okajima, T.; et al. Novel antibacterial compounds specifically targeting the essential WalR response regulator. J. Antibiot. 2010, 63, 127–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivakumar, K.V.; Karunakar, P.; Chatterjee, J. Inhibition of NarL of Mycobacterium tuberculosis: An in silico approach. Interdiscip. Sci. 2014, 6, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ren, J.; Harlos, K.; Jones, D.M.; Zeltina, A.; Bowden, T.A.; Padilla-Parra, S.; Fry, E.E.; Stuart, D.I. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 2016, 535, 169–172. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Wang, Z.; Li, L.; Dong, S.; Li, Z.; Jiang, Z.; Wang, Y.; Shui, W. Novel chemical ligands to Ebola virus and Marburg virus nucleoproteins identified by combining affinity mass spectrometry and metabolomics approaches. Sci. Rep. 2016, 6, 29680. [Google Scholar] [CrossRef]
- Ren, J.; Zhao, Y.; Fry, E.E.; Stuart, D.I. Target identification and mode of action of four chemically divergent drugs against Ebolavirus infection. J. Med. Chem. 2018, 61, 724–733. [Google Scholar] [CrossRef]
- García-Fernández, E.; Medrano, F.J.; Galán, B.; García, J.L. Deciphering the transcriptional regulation of cholesterol catabolic pathway in mycobacteria: Identification of the inducer of KstR repressor. J. Biol. Chem. 2014, 289, 17576–17588. [Google Scholar] [CrossRef] [Green Version]
- Foulkes, D.M.; Byrne, D.P.; Yeung, W.; Shrestha, S.; Bailey, F.P.; Ferries, S.; Eyers, C.E.; Keeshan, K.; Wells, C.; Drewry, D.H.; et al. Covalent inhibitors of EGFR family protein kinases induce degradation of human Tribbles 2 (TRIB2) pseudokinase in cancer cells. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Nnadi, C.I.; Jenkins, M.L.; Gentile, D.R.; Bateman, L.A.; Zaidman, D.; Balius, T.E.; Nomura, D.K.; Burke, J.E.; Shokat, K.M.; London, N. Novel K-Ras G12C switch-II covalent binders destabilize Ras and accelerate nucleotide exchange. J. Chem. Inf. Model. 2018, 58, 464–471. [Google Scholar] [CrossRef]
- Ishizawa, M.; Akagi, D.; Makishima, M. Lithocholic acid is a vitamin D receptor ligand that acts preferentially in the ileum. Int. J. Mol. Sci. 2018, 19, 1975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pols, T.W.H.; Puchner, T.; Korkmaz, H.I.; Vos, M.; Soeters, M.R.; de Vries, C.J.M. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the Vitamin D receptor. PLoS ONE 2017, 12, e0176715. [Google Scholar] [CrossRef]
- Dang, Z.; Jung, K.; Qian, K.; Lee, K.H.; Huang, L.; Chen, C.H. Synthesis of lithocholic acid derivatives as proteasome regulators. ACS Med. Chem. Lett. 2012, 3, 925–930. [Google Scholar] [CrossRef]
- Kovacs, P.; Csonka, T.; Kovacs, T.; Sari, Z.; Ujlaki, G.; Sipos, A.; Karanyi, Z.; Szeocs, D.; Hegedus, C.; Uray, K.; et al. Lithocholic acid, a metabolite of the microbiome, increases oxidative stress in breast cancer. Cancers 2019, 11, 1255. [Google Scholar] [CrossRef] [Green Version]
- Miko, E.; Vida, A.; Kovacs, T.; Ujlaki, G.; Trencsenyi, G.; Marton, J.; Sari, Z.; Kovacs, P.; Boratko, A.; Hujber, Z.; et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef] [PubMed]
- Luu, T.H.; Bard, J.M.; Carbonnelle, D.; Chaillou, C.; Huvelin, J.M.; Bobin-Dubigeon, C.; Nazih, H. Lithocholic bile acid inhibits lipogenesis and induces apoptosis in breast cancer cells. Cell Oncol. 2018, 41, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Kong, Y.; Han, C.; Chen, J.; Hu, L.; Jiang, H.; Shen, X. D-Alanine:D-alanine ligase as a new target for the flavonoids quercetin and apigenin. Int. J. Antimicrob. Agents 2008, 32, 421–426. [Google Scholar] [CrossRef]
- Zhang, L.; Kong, Y.; Wu, D.; Zhang, H.; Wu, J.; Chen, J.; Ding, J.; Hu, L.; Jiang, H.; Shen, X. Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: Crystal structure characterization with enzymatic inhibition assay. Protein Sci. 2008, 17, 1971–1978. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.H.; Lee, J.H.; Kim, K.T.; Park, Y.S.; Nah, S.Y.; Ahn, D.U.; Paik, H.D. Antimicrobial effect of 7-O-butylnaringenin, a novel flavonoid, and various natural flavonoids against Helicobacter pylori strains. Int. J. Environ. Res. Public Health 2013, 10, 5459–5469. [Google Scholar] [CrossRef]
- Hosoda, K.; Shimomura, H.; Hayashi, S.; Yokota, K.; Hirai, Y. Steroid hormones as bactericidal agents to Helicobacter pylori. FEMS Microbiol. Lett. 2011, 318, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Itoh, M.; Wada, K.; Tan, S.; Kitano, Y.; Kai, J.; Makino, I. Antibacterial action of bile acids against Helicobacter pylori and changes in its ultrastructural morphology: Effect of unconjugated dihydroxy bile acid. J. Gastroenterol. 1999, 34, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, P.G.; Lemos, T.L.; Almeida, M.C.; de Souza, J.M.; Bizerra, A.M.; Santiago, G.M.; da Costa, J.G.; Coutinho, H.D. Lithocholic acid and derivatives: Antibacterial activity. Steroids 2015, 104, 8–15. [Google Scholar] [CrossRef] [PubMed]



| Compound | ΔTm (°C) a | Therapeutic Group b | Chemical Structure |
|---|---|---|---|
| Lorglumide sodium salt | −1.6 | cholecystokinin antagonist |  |
| Meclofenamic acid sodium salt monohydrate | −1.7 | anti-inflammatory, antipyretic |  |
| Miconazole | −2.0 | antifungal |  |
| Tiratricol | −2.3 | hypocholesterolemic, antityroidic hormone |  |
| Propidium iodide | −2.7 | antibacterial |  |
| Diethylstilbestrol | −2.9 | estrogen, antineoplastic |  |
| Dequalinium dichloride | −3.0 | antibacterial |  |
| Lithocholic acid | −3.1 | cholagogue, choleretic |  |
| Simvastatin | −3.5 | antihyperlipidemic |  |
| Dienestrol | −6.8 | non-steroidal estrogen |  |
| ArsR Inhibitor | ITC a | |||
|---|---|---|---|---|
| n | Kd(µM) | ΔH(kcal/mol) | ΔG(kcal/mol) | |
| Tiratricol | 0.74 | 8.6 | −14.4 | −6.9 |
| Propidium iodide | 0.92 | 18 | −19.1 | −6.5 |
| Lithocholic acid | 0.69 | 4.9 | −7.8 | −7.2 |
| Lorglumide sodium salt | 0.78 | 0.67 | 1.8 | −8.4 |
| Drug | MIC (MBC), mg/L | ||
|---|---|---|---|
| ATCC 700392 | ATCC 43504 (MTZ-R) | ATCC 700684 (CLR-R) | |
| Tiratricol | 64 (64) | 64 (64) | 64 (64) |
| Propidium iodide | 16 (32) | 16 (32) | 16 (32) |
| Lithocholic acid | 32 (32) | 32 (32) | 32 (32) |
| Lorglumide | 64 (64) | 64 (64) | 64 (128) |
| Metronidazole | 1 (2) | 64 (128) | 1 (2) |
| Clarithromycin | ≤ 0.12 (≤ 0.12) | ≤ 0.12 (≤ 0.12) | 64 (128) |
| Strain | Combination Tested | FICdrug | FICLCA | FICI a | Interaction b |
|---|---|---|---|---|---|
| ATCC 700684 | LCA + Clarithromycin | 0.031 | 0.5 | 0.53 | Additive |
| ATCC 43504 | LCA + Metronidazole | 1 | 1 | 2 | No interaction |
| ATCC 700392 | LCA + Levofloxacin | 0.5 | 0.5 | 1 | Additive |
| ATCC 700392 | LCA + Chrysin | 0.25 | 0.5 | 0.75 | Additive |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, A.; Casado, J.; Chueca, E.; Salillas, S.; Velázquez-Campoy, A.; Sancho, J.; Lanas, Á. Small Molecule Inhibitors of the Response Regulator ArsR Exhibit Bactericidal Activity against Helicobacter pylori. Microorganisms 2020, 8, 503. https://doi.org/10.3390/microorganisms8040503
González A, Casado J, Chueca E, Salillas S, Velázquez-Campoy A, Sancho J, Lanas Á. Small Molecule Inhibitors of the Response Regulator ArsR Exhibit Bactericidal Activity against Helicobacter pylori. Microorganisms. 2020; 8(4):503. https://doi.org/10.3390/microorganisms8040503
Chicago/Turabian StyleGonzález, Andrés, Javier Casado, Eduardo Chueca, Sandra Salillas, Adrián Velázquez-Campoy, Javier Sancho, and Ángel Lanas. 2020. "Small Molecule Inhibitors of the Response Regulator ArsR Exhibit Bactericidal Activity against Helicobacter pylori" Microorganisms 8, no. 4: 503. https://doi.org/10.3390/microorganisms8040503