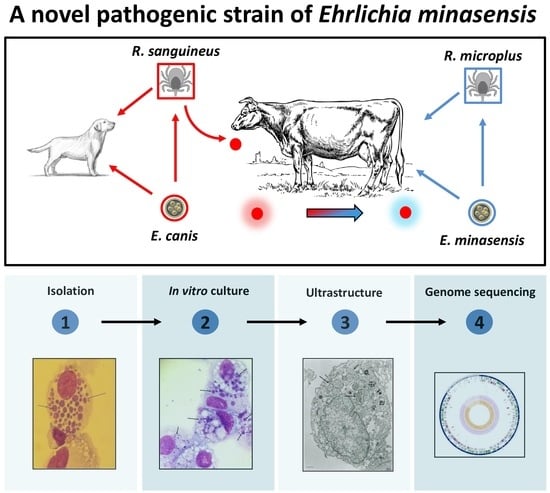
Isolation and Characterization of a Novel Pathogenic Strain of Ehrlichia minasensis
Abstract
:1. Introduction
2. Material and Methods
2.1. Animal and Sample Collection
2.2. DNA Extraction and Polymerase Chain Reaction
2.3. In Vitro Culture of Ehrlichia
2.4. Transmission Electron Microscopy
2.5. Genome Sequencing
2.6. Core-Genome-Based Phylogenetic Tree
2.7. Bacterial Pan-Genome Profiles
2.8. Genome Comparative Analysis
3. Results
3.1. Natural Infection of E. minasensis in a Calf
3.2. Isolation and In Vitro Culture of E. minasensis
3.3. Electron Microscopy Characterization
3.4. Genome Properties and Phylogenetic Analysis
3.5. Genome Comparison Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dumler, J.S.; Barbet, A.F.; Bekker, C.P.; Dasch, G.A.; Palmer, G.H.; Ray, S.C.; Rikihisa, Y.; Rurangirwa, F.R. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: Unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001, 51, 2145–2165. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Cruz, A.; Zweygarth, E.; Vancová, M.; Broniszewska, M.; Grubhoffer, L.; Passos, L.M.F.; Ribeiro, M.F.B.; Alberdi, P.; de la Fuente, J. Ehrlichia minasensis sp. nov., isolated from the tick Rhipicephalus microplus. Int. J. Syst. Evol. Microbiol. 2016, 66, 1426–1430. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, B.A. Heartwater—Ehrlichia ruminantium infection. Rev. Sci. Tech. 2015, 34, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Gondard, M.; Cabezas-Cruz, A.; Charles, R.A.; Vayssier-Taussat, M.; Albina, E.; Moutailler, S. Ticks and Tick-Borne Pathogens of the Caribbean: Current Understanding and Future Directions for More Comprehensive Surveillance. Front. Cell. Infect. Microbiol. 2017, 7, 490. [Google Scholar] [CrossRef]
- Gajadhar, A.A.; Lobanov, V.; Scandrett, W.B.; Campbell, J.; Al-Adhami, B. A novel Ehrlichia genotype detected in naturally infected cattle in North America. Vet. Parasitol. 2010, 173, 324–329. [Google Scholar] [CrossRef]
- Lobanov, V.A.; Gajadhar, A.A.; Al-Adhami, B.; Schwantje, H.M. Molecular study of free-ranging mule deer and white-tailed deer from British Columbia, Canada, for evidence of Anaplasma spp. and Ehrlichia spp. Transbound. Emerg. Dis. 2012, 59, 233–243. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Zweygarth, E.; Ribeiro, M.F.; da Silveira, J.A.; de la Fuente, J.; Grubhoffer, L.; Valdés, J.J.; Passos, L.M. New species of Ehrlichia isolated from Rhipicephalus (Boophilus) microplus shows an ortholog of the E. canis major immunogenic glycoprotein gp36 with a new sequence of tandem repeats. Parasit. Vectors 2012, 5, 291. [Google Scholar] [CrossRef]
- Zweygarth, E.; Schöl, H.; Lis, K.; Cabezas-Cruz, A.; Thiel, C.; Silaghi, C.; Ribeiro, M.F.; Passos, L.M. In vitro culture of a novel genotype of Ehrlichia sp. from Brazil. Transbound. Emerg. Dis. 2013, 60, 86–92. [Google Scholar] [CrossRef]
- Aguiar, D.M.; Ziliani, T.F.; Zhang, X.; Melo, A.L.; Braga, I.A.; Witter, R.; Freitas, L.C.; Rondelli, A.L.; Luis, M.A.; Sorte, E.C.; et al. A novel Ehrlichia genotype strain distinguished by the TRP36 gene naturally infects cattle in Brazil and causes clinical manifestations associated with ehrlichiosis. Ticks Tick Borne Dis. 2014, 5, 537–544. [Google Scholar] [CrossRef]
- Carvalho, I.T.S.; Melo, A.L.T.; Freitas, L.C.; Verçoza, R.V.; Alves, A.S.; Costa, J.S.; Chitarra, C.S.; Nakazato, L.; Dutra, V.; Pacheco, R.C.; et al. Minimum infection rate of Ehrlichia minasensis in Rhipicephalus microplus and Amblyomma sculptum ticks in Brazil. Ticks Tick. Dis. 2016, 7, 849–852. [Google Scholar] [CrossRef]
- Iweriebor, B.C.; Mmbaga, E.J.; Adegborioye, A.; Igwaran, A.; Obi, L.C.; Okoh, A.I. Genetic profiling for Anaplasma and Ehrlichia species in ticks collected in the Eastern Cape Province of South Africa. BMC Microbiol. 2017, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Cicculli, V.; Masse, S.; Capai, L.; de Lamballerie, X.; Charrel, R.; Falchi, A. First detection of Ehrlichia minasensis in Hyalomma marginatum ticks collected from cattle in Corsica, France. Vet. Med. Sci. 2019, 5, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Conraths, F.J.; Sauter-Louis, C.; Krücken, J.; Nijhof, A.M. Epidemiology of tick-borne pathogens in the semi-arid and the arid agro-ecological zones of Punjab province, Pakistan. Transbound. Emerg. Dis. 2019, 66, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, X.; Mu, J.; Yu, X.; Fei, Y.; Chang, J.; Bi, Y.; Zhou, Y.; Ding, Z.; Yin, R. Emergence of a Novel Ehrlichia minasensis Strain, Harboring the Major Immunogenic Glycoprotein trp36 with Unique Tandem Repeat and C-Terminal Region Sequences, in Haemaphysalis hystricis Ticks Removed from Free-Ranging Sheep in Hainan Province, China. Microorganisms 2019, 7, 369. [Google Scholar] [CrossRef]
- Koh, F.X.; Kho, K.L.; Kisomi, M.G.; Wong, L.P.; Bulgiba, A.; Tan, P.E.; Lim, Y.A.L.; Nizam, Q.N.H.; Panchadcharam, C.; Tay, S.T. Ehrlichia and Anaplasma Infections: Serological Evidence and Tick Surveillance in Peninsular Malaysia. J. Med. Entomol. 2018, 55, 269–276. [Google Scholar] [CrossRef]
- Hailemariam, Z.; Krücken, J.; Baumann, M.; Ahmed, J.S.; Clausen, P.H.; Nijhof, A.M. Molecular detection of tick-borne pathogens in cattle from Southwestern Ethiopia. PLoS ONE 2017, 12, e0188248. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Zweygarth, E.; Aguiar, D.M. Ehrlichia minasensis, an old demon with a new name. Ticks Tick Borne Dis. 2019, 10, 828–829. [Google Scholar] [CrossRef]
- Meyer, D.J.; Harvey, J.W. Veterinary Laboratory Medicine: Interpretation & Diagnosis; Saunders: Philadelphia, PA, USA, 2004; ISBN 0721689264. [Google Scholar]
- Aguiar, D.M.; Hagiwara, M.K.; Labruna, M.B. In vitro isolation and molecular characterization of an Ehrlichia canis strain from São Paulo, Brazil. Braz. J. Microbiol. 2008, 39, 489–493. [Google Scholar] [CrossRef]
- Popov, V.L.; Han, V.C.; Chen, S.M.; Dumler, J.S.; Feng, H.M.; Andreadis, T.G.; Tesh, R.B.; Walker, D.H. Ultrastructural differentiation of the genogroups in the genus Ehrlichia. J. Med. Microbiol. 1998, 47, 235–251. [Google Scholar] [CrossRef]
- Aguiar, D.M.; Araujo, J.P., Jr.; Nakazato, L.; Bard, E.; Cabezas-Cruz, A. Complete genome sequence of an Ehrlichia minasensis strain isolated from cattle. Microbiol. Resour. Announc. 2019, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranwez, V.; Harispe, S.; Delsuc, F.; Douzery, E.J.P. MACSE: Multiple Alignment of Coding Sequences Accounting for Frameshifts and Stop Codons. PLoS ONE 2011, 6, e22594. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Zhao, Y.; Jia, X.; Yang, J.; Ling, Y.; Zhang, Z.; Wu, J.Y.J.; Xiao, J. PanGP: A tool for quickly analyzing bacterial pan-genome profile. Bioinformatics 2014, 30, 1297–1299. [Google Scholar] [CrossRef] [Green Version]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 5 September 2018).
- Aguiar, D.M. Ehrlichiosis. In Emerging and Re-emerging Infectious Diseases of Livestock; Bayry, J., Ed.; Springer: Cham, Switzerland, 2017; pp. 365–375. [Google Scholar] [CrossRef]
- Harrus, S.; Waner, T.; Neer, T.M. Ehrlichia canis infection. In Infectious Diseases of the Dog and Cat; Greene, C.E., Ed.; Elsevier: St. Louis, MO, USA, 2012; ISBN 9781416061304. [Google Scholar]
- Al-Adhami, B.; Scandrett, W.B.; Lobanov, V.A.; Gajadhar, A.A. Serological cross- reactivity between Anaplasma marginale and an Ehrlichia species in naturally and experimentally infected cattle. J. Vet. Diagn. Investig. 2011, 23, 1181–1188. [Google Scholar] [CrossRef]
- Cabezas-Cruz, A.; Vancová, M.; Zweygarth, E.; Ribeiro, M.F.; Grubhoffer, L.; Passos, L.M. Ultrastructure of Ehrlichia mineirensis, a new member of the Ehrlichia genus. Vet. Microbiol. 2013, 167, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Cruz, A.; Valdés, J.J.; de la Fuente, J. The glycoprotein TRP36 of Ehrlichia sp. UFMG-EV and related cattle pathogen Ehrlichia sp. UFMT-BV evolved from a highly variable clade of E. canis under adaptive diversifying selection. Parasit Vectors 2014, 7, 584. [Google Scholar] [CrossRef] [PubMed]
- De Fine, L.H.H. Does pathogen plasticity facilitate host shifts? PLoS Pathog. 2018, 14, e1006961. [Google Scholar] [CrossRef] [PubMed]




| Features * | Bacteria | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EMI | ECA | ECH | EMU | ER | AC | AM | AP | RC | RP | |
| Size (bp) | 1,335,478 | 1,315,030 | 1,176,248 | 1,196,717 | 1,516,355 | 1,206,806 | 1,136,981 | 1,477,581 | 1,268,755 | 1,111,612 |
| GC (%) | 29.5 | 29.0 | 30.1 | 29.7 | 27.5 | 50.0 | 49.8 | 42.0 | 32.4 | 29.0 |
| tRNA | 36 | 36 | 37 | 37 | 36 | 37 | 35 | 37 | 33 | 33 |
| rRNA | 3 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 2 |
| total ORFs | 1270 | 1068 | 1032 | 1055 | 1056 | 1194 | 1359 | 1501 | 1637 | 960 |
| PATRIC CDS | 1231 | 1030 | 992 | 992 | 1015 | 1153 | 1321 | 1447 | 1578 | 919 |
| Contigs | 55 | 1 | 1 | 1 | 1 | 1 | 204 | 1 | 1 | 1 |
| Class | EMI | ECA | ECH | EMU | ER | AC | AM | AP | RC | RP |
|---|---|---|---|---|---|---|---|---|---|---|
| Amino Acids and Derivatives | 0.034 (42) | 0.036 (37) | 0.037 (37) | 0.041 (41) | 0.042 (43) | 0.021 (24) | 0.030 (39) | 0.005*** (7) | 0.015** (24) | 0.026 (24) |
| Carbohydrates | 0.002 (2) | 0.002 (2) | 0.002 (2) | 0.002 (2) | 0.002 (2) | 0.002 (2) | 0.002 (2) | 0.001 (2) | 0.001 (2) | 0.002 (2) |
| Cell Cycle, Cell Division and Death | 0.011 (13) | 0.006 (6) | 0.006 (6) | 0.006 (6) | 0.006 (6) | 0.029** (33) | 0.036*** (47) | 0.004 (6) | 0.029** (45) | 0.032*** (29) |
| Cell Envelope, Capsule and Slime layer | 0.002 (2) | 0.002 (2) | 0.002 (2) | 0.002 (2) | 0.002 (2) | 0.004 (5) | 0.004 (5) | 0.001 (2) | 0.008* (13) | 0.014** (13) |
| Clustering-based subsystems | 0.003 (4) | 0.0165** (17) | 0.017** (17) | 0.016** (16) | 0.016** (16) | 0.015** (17) | 0.004 (5) | 0.007 (10) | 0.003 (4) | 0.004 (4) |
| Cofactors, Vitamins, Prosthetic Groups | 0.084 (104) | 0.098 (101) | 0.102 (101) | 0.104 (103) | 0.101 (103) | 0.087 (100) | 0.090 (119) | 0.077 (111) | 0.042*** (66) | 0.069 (63) |
| DNA Processing | 0.032 (40) | 0.039 (40) | 0.040 (40) | 0.042 (42) | 0.044 (45) | 0.042 (49) | 0.048 (63) | 0.036 (52) | 0.034 (54) | 0.038 (35) |
| Energy and Precursor Metabolites Generation | 0.037 (46) | 0.045 (46) | 0.046 (46) | 0.050 (50) | 0.046 (47) | 0.040 (46) | 0.039 (52) | 0.037 (53) | 0.027 (42) | 0.041 (38) |
| Fatty Acids, Lipids, and Isoprenoids | 0.037 (45) | 0.043 (44) | 0.044 (44) | 0.045 (45) | 0.041 (42) | 0.036 (42) | 0.028 (37) | 0.029 (42) | 0.018** (28) | 0.030 (28) |
| Membrane Transport | 0.044 (54) | 0.046 (47) | 0.047 (47) | 0.049 (49) | 0.054 (55) | 0.056 (65) | 0.058 (77) | 0.039 (57) | 0.037 (59) | 0.063 (58) |
| Metabolite damage and its repair or mitigation | 0.003 (4) | 0.004 (4) | 0.004 (4) | 0.004 (4) | 0.004 (4) | 0.003 (4) | 0.003 (4) | 0.003 (4) | 0.003 (4) | 0.003 (3) |
| Miscellaneous | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.002 (2) | 0.002 (2) | 0.001 (1) | 0.000 (0) | 0.001 (1) |
| Nucleosides and Nucleotides | 0.025 (31) | 0.030 (31) | 0.031 (31) | 0.033 (33) | 0.031 (31) | 0.027 (31) | 0.026 (35) | 0.029 (42) | 0.001*** (2) | 0.002 (2) |
| Phosphate Metabolism | 0.003 (4) | 0.004 (4) | 0.004 (4) | 0.004 (4) | 0.004 (4) | 0.003 (4) | 0.005 (7) | 0.003 (4) | 0.000 (0) | 0.000 (0) |
| Prokaryotic cell type differentiation | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) | 0.001 (1) |
| Protein Fate (folding, modification, targeting, degradation) | 0.024 (29) | 0.028 (29) | 0.029 (29) | 0.029 (29) | 0.029 (29) | 0.024 (28) | 0.024 (32) | 0.020 (29) | 0.016 (26) | 0.028 (26) |
| Protein Synthesis | 0.130 (160) | 0.149 (153) | 0.154 (153) | 0.119 (118) | 0.157 (159) | 0.101* (117) | 0.129 (170) | 0.112 (162) | 0.097** (153) | 0.164* (151) |
| Respiration | 0.076 (94) | 0.089 (92) | 0.093 (92) | 0.098 (97) | 0.101* (103) | 0.075*** (87) | 0.074 (98) | 0.068 (99) | 0.059 (93) | 0.108* (99) |
| RNA Processing | 0.030 (37) | 0.028 (29) | 0.029 (29) | 0.034 (34) | 0.031 (31) | 0.025 (29) | 0.030 (40) | 0.020 (29) | 0.032 (50) | 0.051* (47) |
| Stress Response, Defense and Virulence | 0.040 (49) | 0.038 (39) | 0.039 (39) | 0.040 (40) | 0.040 (41) | 0.035 (40) | 0.042 (56) | 0.025* (36) | 0.034 (54) | 0.049 (45) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moura de Aguiar, D.; Pessoa Araújo Junior, J.; Nakazato, L.; Bard, E.; Aguilar-Bultet, L.; Vorimore, F.; Leonidovich Popov, V.; Moleta Colodel, E.; Cabezas-Cruz, A. Isolation and Characterization of a Novel Pathogenic Strain of Ehrlichia minasensis. Microorganisms 2019, 7, 528. https://doi.org/10.3390/microorganisms7110528
Moura de Aguiar D, Pessoa Araújo Junior J, Nakazato L, Bard E, Aguilar-Bultet L, Vorimore F, Leonidovich Popov V, Moleta Colodel E, Cabezas-Cruz A. Isolation and Characterization of a Novel Pathogenic Strain of Ehrlichia minasensis. Microorganisms. 2019; 7(11):528. https://doi.org/10.3390/microorganisms7110528
Chicago/Turabian StyleMoura de Aguiar, Daniel, João Pessoa Araújo Junior, Luciano Nakazato, Emilie Bard, Lisandra Aguilar-Bultet, Fabien Vorimore, Vsevolod Leonidovich Popov, Edson Moleta Colodel, and Alejandro Cabezas-Cruz. 2019. "Isolation and Characterization of a Novel Pathogenic Strain of Ehrlichia minasensis" Microorganisms 7, no. 11: 528. https://doi.org/10.3390/microorganisms7110528