Exo-Metabolites of Phaseolus vulgaris-Nodulating Rhizobial Strains
Abstract
:1. Introduction
2. Results
2.1. Bacterial Growth and Sample Preparation
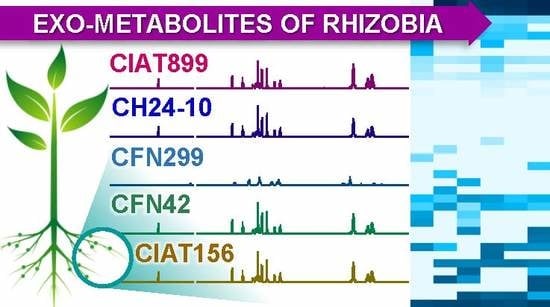
2.2. Exo-Metabolite Identification by NMR
2.3. Statistical Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Growth and Sample Preparation
4.2. Exo-Metabolite Identification by NMR
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mapelli, V.; Olsson, L.; Nielsen, J. Metabolic footprinting in microbiology: Methods and applications in functional genomics and biotechnology. Trends Biotechnol. 2008, 26, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Behrends, V.; Williams, H.D.; Bundy, J.G. Metabolic footprinting: Extracellular metabolomic analysis. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.-L., Eds.; Springer New York: New York, NY, USA, 2014; pp. 281–292. [Google Scholar]
- Palama, T.L.; Canard, I.; Rautureau, G.J.; Mirande, C.; Chatellier, S.; Elena-Herrmann, B. Identification of bacterial species by untargeted NMR spectroscopy of the exo-metabolome. Analyst 2016, 141, 4558–4561. [Google Scholar] [CrossRef]
- Guinel, F.C. Ethylene, a hormone at the center-stage of nodulation. Front. Plant Sci. 2015, 6, 21. [Google Scholar] [CrossRef]
- Peix, A.; Velazquez, E.; Silva, L.R.; Mateos, P.F. Key Molecules Involved in Beneficial Infection Process in Rhizobia-Legume Symbiosis; Springer-Verlag Wien: Vienna, Austria, 2010; pp. 55–80. [Google Scholar]
- Gibson, K.E.; Kobayashi, H.; Walker, G.C. Molecular determinants of a symbiotic chronic infection. In Annual Review of Genetics; Annual Reviews: Palo Alto, CA, USA, 2008; Volume 42, pp. 413–441. [Google Scholar]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291. [Google Scholar] [CrossRef]
- Mitsch, M.J.; diCenzo, G.C.; Cowie, A.; Finan, T.M. Succinate transport is not essential for symbiotic nitrogen fixation by Sinorhizobium meliloti or Rhizobium leguminosarum. Appl. Environ. Microbiol. 2018, 84, 15. [Google Scholar] [CrossRef] [PubMed]
- Janczarek, M. Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 2011, 12, 7898–7933. [Google Scholar] [CrossRef]
- Arora, N.K.; Khare, E.; Singh, S.; Maheshwari, D.K. Effect of Al and heavy metals on enzymes of nitrogen metabolism of fast and slow growing rhizobia under explanta conditions. World J. Microbiol. Biotechnol. 2010, 26, 811–816. [Google Scholar] [CrossRef]
- Keum, Y.S.; Seo, J.S.; Li, Q.X.; Kim, J.H. Comparative metabolomic analysis of Sinorhizobium sp. C4 during the degradation of phenanthrene. Appl. Microbiol. Biotechnol. 2008, 80, 863–872. [Google Scholar] [CrossRef]
- Janczarek, M.; Rachwal, K.; Marzec, A.; Grzadziel, J.; Palusinska-Szysz, M. Signal molecules and cell-surface components involved in early stages of the legume-rhizobium interactions. Appl. Soil Ecol. 2015, 85, 94–113. [Google Scholar] [CrossRef]
- Marczak, M.; Mazur, A.; Koper, P.; Zebracki, K.; Skorupska, A. Synthesis of rhizobial exopolysaccharides and their importance for symbiosis with legume plants. Genes 2017, 8, 24. [Google Scholar] [CrossRef]
- Mia, M.B.; Shamsuddin, Z. Rhizobium as a crop enhancer and biofertilizer for increased cereal production. Afr. J. Biotechnol. 2010, 9, 6001–6009. [Google Scholar]
- Gemperline, E.; Jayaraman, D.; Maeda, J.; Ane, J.M.; Li, L.J. Multifaceted investigation of metabolites during nitrogen fixation in medicago via high resolution MALDI-MS Imaging and ESI-MS. J. Am. Soc. Mass Spectrom. 2015, 26, 149–158. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Martyn, A.; Kopriva, S. Exometabolomic profiling of bacterial strains as cultivated using arabidopsis root extract as the sole carbon source. Mol. Plant Microbe Interact. 2018, 31, 803–813. [Google Scholar] [CrossRef]
- Dear, G.J.; Roberts, A.D.; Beaumont, C.; North, S.E. Evaluation of preparative high performance liquid chromatography and cryoprobe-nuclear magnetic resonance spectroscopy for the early quantitative estimation of drug metabolites in human plasma. J. Chromatogr. B 2008, 876, 182–190. [Google Scholar] [CrossRef]
- Molinski, T.F. Nanomole-scale natural products discovery. Curr. Opin. Drug Discov. Dev. 2009, 12, 197–206. [Google Scholar]
- Tang, J. Microbial metabolomics. Curr. Genom. 2011, 12, 391–403. [Google Scholar] [CrossRef]
- Mashego, M.R.; Rumbold, K.; De Mey, M.; Vandamme, E.; Soetaert, W.; Heijnen, J.J. Microbial metabolomics: Past, present and future methodologies. Biotechnol. Lett. 2007, 29, 1–16. [Google Scholar] [CrossRef]
- Puntus, I.; Sakharovsky, V.; Filonov, A.; Boronin, A. Surface activity and metabolism of hydrocarbon-degrading microorganisms growing on hexadecane and naphthalene. Process Biochem. 2005, 40, 2643–2648. [Google Scholar] [CrossRef]
- Lee, J.-E.; Hwang, G.-S.; Lee, C.-H.; Hong, Y.-S. Metabolomics reveals alterations in both primary and secondary metabolites by wine bacteria. J. Agric. Food Chem. 2009, 57, 10772–10783. [Google Scholar] [CrossRef]
- Murovec, B.; Makuc, D.; Repinc, S.K.; Prevoršek, Z.; Zavec, D.; Šket, R.; Pečnik, K.; Plavec, J.; Stres, B. 1H NMR metabolomics of microbial metabolites in the four MW agricultural biogas plant reactors: A case study of inhibition mirroring the acute rumen acidosis symptoms. J. Environ. Manag. 2018, 222, 428–435. [Google Scholar] [CrossRef]
- Hoerr, V.; Duggan, G.E.; Zbytnuik, L.; Poon, K.K.; Große, C.; Neugebauer, U.; Methling, K.; Löffler, B.; Vogel, H.J. Characterization and prediction of the mechanism of action of antibiotics through NMR metabolomics. BMC Microbiol. 2016, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Sheng, J.; Fu, Y.; Li, M.; Wang, J.; Jia, A.-Q. 1H NMR-based global metabolic studies of pseudomonas aeruginosa upon exposure of the quorum sensing inhibitor resveratrol. J. Proteome Res. 2017, 16, 824–830. [Google Scholar] [CrossRef]
- Griffith, C.M.; Morgan, M.A.; Dinges, M.M.; Mathon, C.; Larive, C.K. Metabolic profiling of chloroacetanilide herbicides in earthworm coelomic fluid using 1H NMR and GC-MS. J. Proteome Res. 2018, 17, 2611–2622. [Google Scholar] [CrossRef] [PubMed]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Baharum, S.N.; Azizan, K.A. Metabolomics in systems biology. In Omics Applications for Systems Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 51–68. [Google Scholar]
- Wishart, D.S. Quantitative metabolomics using NMR. TrAC Trends Anal. Chem. 2008, 27, 228–237. [Google Scholar] [CrossRef]
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Koek, M.M.; Muilwijk, B.; van der Werf, M.J.; Hankemeier, T. Microbial metabolomics with gas chromatography/mass spectrometry. Anal. Chem. 2006, 78, 1272–1281. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Puebla, S.T.; Hernández, M.A.R.; Ruiz, G.G.; Ormeño-Orrillo, E.; Martinez-Romero, J.C.; Servín-Garcidueñas, L.E.; Núñez-de la Mora, A.; Amescua-Villela, G.; Negrete-Yankelevich, S.; Martínez-Romero, E. Nodule bacteria from the cultured legume Phaseolus dumosus (belonging to the Phaseolus vulgaris cross-inoculation group) with common tropici phenotypic characteristics and symbiovar but distinctive phylogenomic position and chromid. Syst. Appl. Microbiol. 2018, in press. [Google Scholar]
- Taylor, D.G.; Trudgill, P.W.; Cripps, R.E.; Harris, P.R. The microbial metabolism of acetone. Microbiology 1980, 118, 159–170. [Google Scholar] [CrossRef]
- Kouchi, H.; Fukai, K.; Kihara, A. Metabolism of glutamate and aspartate in bacteroids isolated from soybean root nodules. Microbiology 1991, 137, 2901–2910. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Jiménez, K.; Sohlenkamp, C.; Geiger, O.; Martínez-Romero, E.; Werner, D.; Vinuesa, P. A ClC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol. Plant Microbe Interact. 2005, 18, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Vences-Guzmán, M.Á.; Guan, Z.; Ormeño-Orrillo, E.; González-Silva, N.; López-Lara, I.M.; Martínez-Romero, E.; Geiger, O.; Sohlenkamp, C. Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol. Microbiol. 2011, 79, 1496–1514. [Google Scholar] [CrossRef] [PubMed]
- Taté, R.; Riccio, A.; Caputo, E.; laccarino, M.; Patriarca, E.J. The Rhizobium etli metZ gene is essential for methionine biosynthesis and nodulation of Phaseolus vulgaris. Mol. Plant Microbe Interact. 1999, 12, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.J.; Heys, R.; Martin, T.; Savard, M. Sinorhizobium meliloti Cells Require Biotin and either Cobalt or Methionine for Growth. Appl. Environ. Microb. 2001, 67, 3767–3770. [Google Scholar] [CrossRef]
- Szilagyi-Zecchin, V.J.; Mógor, Á.F.; Figueiredo, G.G.O. Strategies for characterization of agriculturally important bacteria. In Microbial Inoculants in Sustainable Agricultural Productivity: Vol. 1: Research Perspectives; Singh, D.P., Singh, H.B., Prabha, R., Eds.; Springer India: New Delhi, India, 2016; pp. 1–21. [Google Scholar]
- Barra, L.; Fontenelle, C.; Ermel, G.; Trautwetter, A.; Walker, G.C.; Blanco, C. Interrelations between glycine betaine catabolism and methionine biosynthesis in Sinorhizobium meliloti strain 102F34. J. Bacteriol. 2006, 188, 7195–7204. [Google Scholar] [CrossRef] [PubMed]
- de Rudder, K.E.; Sohlenkamp, C.; Geiger, O. Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 1999, 274, 20011–20016. [Google Scholar] [CrossRef]
- Vauclare, P.; Bligny, R.; Gout, E.; Widmer, F. An overview of the metabolic differences between Bradyrhizobium japonicum 110 bacteria and differentiated bacteroids from soybean (Glycine max) root nodules: An in vitro 13C-and 31P-nuclear magnetic resonance spectroscopy study. FEMS Microbiol. Lett. 2013, 343, 49–56. [Google Scholar] [CrossRef]
- Streeter, J.G. Accumulation of alpha, alpha-trehalose by Rhizobium bacteria and bacteroids. J. Bacteriol. 1985, 164, 78–84. [Google Scholar]
- Farías-Rodríguez, R.; Mellor, R.B.; Arias, C.; Peña-Cabriales, J.J. The accumulation of trehalose in nodules of several cultivars of common bean (Phaseolus vulgaris) and its correlation with resistance to drought stress. Physiol. Plantarum 1998, 102, 353–359. [Google Scholar] [CrossRef]
- McIntyre, H.J.; Davies, H.; Hore, T.A.; Miller, S.H.; Dufour, J.-P.; Ronson, C.W. Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccation tolerance. Appl. Environ. Microbiol. 2007, 73, 3984–3992. [Google Scholar] [CrossRef]
- Wang, C.X.; Saldanha, M.; Sheng, X.Y.; Shelswell, K.J.; Walsh, K.T.; Sobral, B.W.S.; Charles, T.C. Roles of poly-3-hydroxybutyrate (PHB) and glycogen in symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology 2007, 153, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Copeland, L. Acetyl coenzyme A acetyltransferase of Rhizobium sp. (Cicer) strain CC 1192. Appl. Environ. Microb. 1997, 63, 3432–3437. [Google Scholar]
- Obruca, S.; Sedlacek, P.; Krzyzanek, V.; Mravec, F.; Hrubanova, K.; Samek, O.; Kucera, D.; Benesova, P.; Marova, I. Accumulation of poly (3-hydroxybutyrate) helps bacterial cells to survive freezing. PLoS ONE 2016, 11, e0157778. [Google Scholar] [CrossRef]
- Korneyeva, M.; Hotta, J.; Lebing, W.; Rosenthal, R.; Franks, L.; Petteway, S., Jr. Enveloped virus inactivation by caprylate: A robust alternative to solvent-detergent treatment in plasma derived intermediates. Biologicals 2002, 30, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Alsaeed, T.I. Antibacterial eFFICacy and Discoloration Effect of a Novel Intracanal Antibiotic Dressing. Ph.D. Thesis, University of Maryland, Baltimore, MD, USA, 2015. [Google Scholar]
- Angenent, L.T.; Kucek, L. Methods of and Systems for Producing Caprylic Acid And/Or Caprylate. US Patents 15/607,188, 30 November 2017. [Google Scholar]
- Reddy, M.V.; Mohan, S.V.; Chang, Y.-C. Medium-Chain Fatty Acids (MCFA) Production Through Anaerobic Fermentation Using Clostridium kluyveri: Effect of Ethanol and Acetate. Appl. Biochem. Biotech. 2018, 185, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Van Schie, B.; De Mooy, O.; Linton, J.; Van Dijken, J.; Kuenen, J. PQQ-dependent production of gluconic acid by Acinetobacter, Agrobacterium and Rhizobium species. Microbiology 1987, 133, 867–875. [Google Scholar] [CrossRef]
- Giles, C.D.; Hsu, P.-C.; Richardson, A.E.; Hurst, M.R.; Hill, J.E. The role of gluconate production by Pseudomonas spp. in the mineralization and bioavailability of calcium–phytate to Nicotiana tabacum. Can. J. Microbiol. 2015, 61, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Skorupska, A.; Janczarek, M.; Marczak, M.; Mazur, A.; Król, J. Rhizobial exopolysaccharides: Genetic control and symbiotic functions. Microb. Cell Fact. 2006, 5, 7. [Google Scholar] [CrossRef]
- Sjöberg, A.; Hahn-Hägerdal, B. β-Glucose-1-phosphate, a possible mediator for polysaccharide formation in maltose-assimilating Lactococcus lactis. Appl. Environ. Microb. 1989, 55, 1549–1554. [Google Scholar]
- Kim, Y.-S. Malonate metabolism: Biochemistry, molecular biology, physiology, and industrial application. BMB Rep. 2002, 35, 443–451. [Google Scholar] [CrossRef]
- An, J.H.; Lee, H.Y.; Ko, K.N.; Kim, E.-S.; Kim, Y.S. Symbiotic effects of deltamatB Rhizobium leguminosarum bv. trifolii mutant on clovers. Mol. Cells 2002, 14, 261–266. [Google Scholar] [PubMed]
- Vogels, G.v.d.; Van der Drift, C. Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 1976, 40, 403. [Google Scholar]
- Tanaka, Y.; Onaka, T.; Matsui, T.; Maruhashi, K.; Kurane, R. Desulfurization of benzothiophene by the gram-negative bacterium, Sinorhizobium sp. KT55. Curr. Microbiol. 2001, 43, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Stowers, M.D. Carbon metabolism in Rhizobium species. Ann. Rev. Microbiol. 1985, 39, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Trinchant, J.C.; Rigaud, J. Lactate dehydrogenase from Rhizobium. purification and role in indole metabolism. Physiol. Plantarum 1974, 32, 394–399. [Google Scholar] [CrossRef]
- El-Sayed, A.K.; Hothersall, J.; Cooper, S.M.; Stephens, E.; Simpson, T.J.; Thomas, C.M. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 2003, 10, 419–430. [Google Scholar] [CrossRef]
- Hopwood, D.A.; Sherman, D.H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 1990, 24, 37–62. [Google Scholar] [CrossRef]
- Moorhouse, P.C.; Grootveld, M.; Halliwell, B.; Quinlan, J.G.; Gutteridge, J.M. Allopurinol and oxypurinol are hydroxyl radical scavengers. FEBS Lett. 1987, 213, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Atkins, C.A.; Sanford, P.J.; Storer, P.J.; Pate, J.S. Inhibition of nodule functioning in cowpea by a xanthine oxidoreductase inhibitor, allopurinol. Plant Physiol. 1988, 88, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Böhringer, J.; Fischer, D.; Mosler, G.; Hengge-Aronis, R. UDP-glucose is a potential intracellular signal molecule in the control of expression of sigma S and sigma S-dependent genes in Escherichia coli. J. Bacteriol. 1995, 177, 413–422. [Google Scholar] [CrossRef]
- Recourt, K.; van Tunen, A.J.; Mur, L.A.; van Brussel, A.A.; Lugtenberg, B.J.; Kijne, J.W. Activation of flavonoid biosynthesis in roots of Vicia sativa subsp. nigra plants by inoculation with Rhizobium leguminosarum biovar viciae. Plant Mol. Biol. 1992, 19, 411–420. [Google Scholar]
- Santos, C.N.S.; Stephanopoulos, G. Melanin-based high-throughput screen for L-tyrosine production in Escherichia coli. Appl. Environ. Microb. 2008, 74, 1190–1197. [Google Scholar] [CrossRef]
- Cubo, M.T.; Buendia-Claveria, A.M.; Beringer, J.E.; Ruiz-Sainz, J.E. Melanin production by Rhizobium strains. Appl. Environ. Microb. 1988, 54, 1812–1817. [Google Scholar]
- Boels, I.C.; Ramos, A.; Kleerebezem, M.; de Vos, W.M. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microb. 2001, 67, 3033–3040. [Google Scholar] [CrossRef]
- Audy, J.; Labrie, S.; Roy, D.; LaPointe, G. Sugar source modulates exopolysaccharide biosynthesis in Bifidobacterium longum subsp. longum CRC 002. Microbiology 2010, 156, 653–664. [Google Scholar] [PubMed]
- Bourque, D.; Pomerleau, Y.; Groleau, D. High-cell-density production of poly-β-hydroxybutyrate (PHB) from methanol by Methylobacterium extorquens: Production of high-molecular-mass PHB. Appl. Microbiol. Biotechnol. 1995, 44, 367–376. [Google Scholar] [CrossRef]
- Le Guennec, A.; Tayyari, F.; Edison, A.S. Alternatives to nuclear overhauser enhancement spectroscopy presat and carr–purcell–meiboom–gill presat for NMR-based metabolomics. Anal. Chem. 2017, 89, 8582–8588. [Google Scholar] [CrossRef]
- Warnes, G.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A. Gplots: Various R Programming Tools for Plotting Data. 2016. (R package version 3.0.). Available online: https://rdrr.io/cran/gplots/ (accessed on 20 October 2018).
- Team, R.C. R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 20 October 2018).




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montes-Grajales, D.; Esturau-Escofet, N.; Esquivel, B.; Martinez-Romero, E. Exo-Metabolites of Phaseolus vulgaris-Nodulating Rhizobial Strains. Metabolites 2019, 9, 105. https://doi.org/10.3390/metabo9060105
Montes-Grajales D, Esturau-Escofet N, Esquivel B, Martinez-Romero E. Exo-Metabolites of Phaseolus vulgaris-Nodulating Rhizobial Strains. Metabolites. 2019; 9(6):105. https://doi.org/10.3390/metabo9060105
Chicago/Turabian StyleMontes-Grajales, Diana, Nuria Esturau-Escofet, Baldomero Esquivel, and Esperanza Martinez-Romero. 2019. "Exo-Metabolites of Phaseolus vulgaris-Nodulating Rhizobial Strains" Metabolites 9, no. 6: 105. https://doi.org/10.3390/metabo9060105