Phage Display to Augment Biomaterial Function
Abstract
:1. Introduction
1.1. Phage Display
1.2. In Vivo Phage Display
1.3. Processing Phage Display Data
1.4. Limitations and Improvements to Phage Display Technology
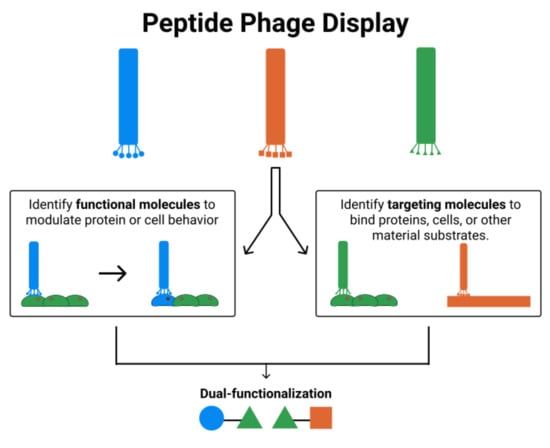
2. Phage Display and Biomaterials
2.1. Functional Molecules
2.1.1. Adhesion and Migration
2.1.2. Differentiation
2.1.3. Inhibition of Cell Function
3. Targeting Molecules
3.1. Locate Specific Cells or Tissue Types
3.1.1. Imaging and Diagnosis
3.1.2. Targeted Drug Delivery
3.2. Material Specific Binding
3.2.1. Silicon
3.2.2. Platinum and Palladium
3.2.3. Apatite
4. Phage Display for Dual-Functioning Peptides
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, S.S. Phage display in pharmaceutical biotechnology. In Current Opinion in Biotechnology; Elsevier Ltd.: Amsterdam, The Netherlands, 2000; pp. 610–616. [Google Scholar] [CrossRef]
- Kehoe, J.W.; Kay, B.K. Filamentous phage display in the new millennium. Chem. Rev. 2005, 105, 4056–4072. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.R.; Deutscher, S.L. In Vivo bacteriophage display for the discovery of novel peptide-based tumor-targeting agents. Methods Mol. Biol. 2009, 504, 275–290. [Google Scholar] [CrossRef]
- Rosenberg, A.; Griffin, K.; Studier, E.W.; Mccormick, M.; Berg, J.; Novy, R.; Mierendorf, R. T7 Select phage display system: A powerful new protein display system based on bacteriophage T7. Innovations 1996, 6, 1–6. [Google Scholar]
- Krumpe, L.R.H.; Atkinson, A.J.; Smythers, G.W.; Kandel, A.; Schumacher, K.M.; McMahon, J.B.; Makowski, L.; Mori, T. T7 lytic phage-displayed peptide libraries exhibit less sequence bias than M13 filamentous phage-displayed peptide libraries. Proteomics 2006, 6, 4210–4222. [Google Scholar] [CrossRef]
- Segvich, S.; Kohn, D.H. Phage display as a strategy for designing organic/inorganic biomaterials. In Biological Interactions on Materials Surfaces; Springer: New York, NY, USA, 2009; pp. 115–132. [Google Scholar] [CrossRef]
- Pasqualini, R.; Ruoslahti, E. Organ targeting in vivo using phage display peptide libraries. Nature 1996, 380, 364–366. [Google Scholar] [CrossRef]
- Wada, A.; Terashima, T.; Kageyama, S.; Yoshida, T.; Narita, M.; Kawauchi, A.; Kojima, H. Efficient prostate cancer therapy with tissue-specific homing peptides identified by advanced phage display technology. Mol. Ther. Oncolytics 2019, 12, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.J.; Joshi, B.P.; Feng, Y.; Gaustad, A.; Fearon, E.R. In vivo fluorescence-based endoscopic detection of colon dysplasia in the mouse using a novel peptide probe. PLoS ONE 2011, 6, e17384. [Google Scholar] [CrossRef] [Green Version]
- Scodeller, P.; Simón-Gracia, L.; Kopanchuk, S.; Tobi, A.; Kilk, K.; Säälik, P.; Kurm, K.; Squadrito, M.L.; Kotamraju, V.R.; Rinken, A.; et al. Precision targeting of tumor macrophages with a CD206 binding peptide. Sci. Rep. 2017, 7, 14655. [Google Scholar] [CrossRef] [Green Version]
- Mann, A.P.; Scodeller, P.; Hussain, S.; Joo, J.; Kwon, E.; Braun, G.B.; Mölder, T.; She, Z.G.; Kotamraju, V.R.; Ranscht, B.; et al. A peptide for targeted, systemic delivery of imaging and therapeutic compounds into acute brain injuries. Nat. Commun. 2016, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Matochko, W.L.; Chu, K.; Jin, B.; Lee, S.W.; Whitesides, G.M.; Derda, R. Deep sequencing analysis of phage libraries using illumina platform. Methods 2012, 58, 47–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, M.R.; Benoit, D.S.W. In vivo translation of peptide-targeted drug delivery systems discovered by phage display. Bioconjugate Chem. 2018, 2161–2169. [Google Scholar] [CrossRef] [PubMed]
- Dias-Neto, E.; Nunes, D.N.; Giordano, R.J.; Sun, J.; Botz, G.H.; Yang, K.; Setubal, J.C.; Pasqualini, R.; Arap, W. Next-generation phage display: Integrating and comparing available molecular tools to enable costeffective high-throughput analysis. PLoS ONE 2009, 4, e8338. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.W.; Yang, L.; De Plano, L.M.; Stackhouse, M.A.; Petrenko, V.A. Evolution of a landscape phage library in a mouse xenograft model of human breast cancer. Viruses 2019, 11, 988. [Google Scholar] [CrossRef] [Green Version]
- Hyde-Deruyscher, R.; Paige, L.A.; Christensen, D.J.; Hyde-Deruyscher, N.; Lim, A.; Fredericks, Z.L.; Kranz, J.; Gallant, P.; Zhang, J.; Rocklage, S.M.; et al. Detection of small-molecule enzyme inhibitors with peptides isolated from phage-displayed combinatorial peptide libraries. Chem. Biol. 2000, 7, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.J.; Ryu, M.Y.; Park, J.P. Identification of high affinity peptides for capturing norovirus capsid proteins. RSC Adv. 2015, 5, 55300–55302. [Google Scholar] [CrossRef]
- Yeh, C.Y.; Hsiao, J.K.; Wang, Y.P.; Lan, C.H.; Wu, H.C. Peptide-conjugated nanoparticles for targeted imaging and therapy of prostate cancer. Biomaterials 2016, 99, 1–15. [Google Scholar] [CrossRef]
- Hwang, H.J.; Ryu, M.Y.; Park, C.Y.; Ahn, J.; Park, H.G.; Choi, C.; Ha, S.-D.; Park, T.J.; Park, J.P. High sensitive and selective electrochemical biosensor: Label-free detection of human norovirus using affinity peptide as molecular binder. Biosens. Bioelectron. 2017, 87, 164–170. [Google Scholar] [CrossRef]
- Huang, X.; Zauscher, S.; Klitzman, B.; Truskey, G.A.; Reichert, W.M.; Kenan, D.J.; Grinstaff, M.W. Peptide interfacial biomaterials improve endothelial cell adhesion and spreading on synthetic polyglycolic acid materials. Ann. Biomed. Eng. 2010, 38, 1965–1976. [Google Scholar] [CrossRef] [Green Version]
- Xing, L.; Xu, Y.; Sun, K.; Wang, H.; Zhang, F.; Zhou, Z.; Zhang, J.; Zhang, F.; Caliskan, B.; Qiu, Z.; et al. Identification of a peptide for folate receptor alpha by phage display and its tumor targeting activity in ovary cancer xenograft. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Segvich, S.; Biswas, S.; Becker, U.; Kohn, D.H. Identification of peptides with targeted adhesion to bone-like mineral via phage display and computational modeling. Cells Tissues Organs 2008, 189, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Peng, J.; He, J.; Li, Q.; Zhou, J.; Liang, X.; Tang, S. Selection and identification of novel peptides specifically targeting human cervical cancer. Amino Acids 2018, 50, 577–592. [Google Scholar] [CrossRef] [PubMed]
- Caprini, A.; Silva, D.; Zanoni, I.; Cunha, C.; Volontè, C.; Vescovi, A.; Gelain, F. A novel bioactive peptide: Assessing its activity over murine neural stem cells and its potential for neural Tissue engineering. N. Biotechnol. 2013, 30, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, J.; Nam, H.K.; Ramaraju, H.; Hatch, N.E.; Kohn, D.H. Inhibition of osteoblast mineralization by phosphorylated phage-derived apatite-specific peptide. Biomaterials 2015, 73, 120–130. [Google Scholar] [CrossRef] [Green Version]
- Rangel, R.; Guzman-Rojas, L.; Le Roux, L.G.; Staquicini, F.I.; Hosoya, H.; Barbu, E.M.; Ozawa, M.G.; Nie, J.; Dunner, K.; Langley, R.R.; et al. Combinatorial targeting and discovery of ligand-Receptors in organelles of mammalian cells. Nat. Commun. 2012, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ryvkin, A.; Ashkenazy, H.; Smelyanski, L.; Kaplan, G.; Penn, O.; Weiss-Ottolenghi, Y.; Privman, E.; Ngam, P.B.; Woodward, J.E.; May, G.D.; et al. Deep panning: Steps towards probing the Igome. PLoS ONE 2012, 7, e41469. [Google Scholar] [CrossRef]
- Ryvkin, A.; Ashkenazy, H.; Weiss-Ottolenghi, Y.; Piller, C.; Pupko, T.; Gershoni, J.M. Phage display peptide libraries: Deviations from randomness and correctives. Nucleic Acids Res. 2018, 46, e52. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Ru, B.; Li, S.; Lin, H.; Guo, F.-B. Sarotup: Scanner and reporter of target-unrelated peptides. BioMed Res. Int. 2010. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Chen, H.; Li, N.; Huang, J. Sarotup: A suite of tools for finding potential target-unrelated peptides from phage display data. Int. J. Biol. Sci. 2019, 15, 1452–1459. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, Y.; Teesalu, T.; Sugahara, K.N.; Kotamrajua, V.R.; Adams, J.D.; Ferguson, B.S.; Gong, Q.; Oh, S.S.; Csordas, A.T.; et al. Selection of phage-displayed peptides on live adherent cells in microfluidic channels. Proc. Natl. Acad. Sci. USA 2011, 108, 6909–6914. [Google Scholar] [CrossRef] [Green Version]
- Matochko, W.L.; Cory Li, S.; Tang, S.K.Y.; Derda, R. Prospective identification of parasitic sequences in phage display screens. Nucleic Acids Res. 2014, 42, 1784–1798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Addison, W.N.; Miller, S.J.; Ramaswamy, J.; Mansouri, A.; Kohn, D.H.; McKee, M.D. Phosphorylation-dependent mineral-type specificity for apatite-binding peptide sequences. Biomaterials 2010, 31, 9422–9430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ’T Hoen, P.A.C.; Jirka, S.M.G.; Ten Broeke, B.R.; Schultes, E.A.; Aguilera, B.; Pang, K.H.; Heemskerk, H.; Aartsma-Rus, A.; Van Ommen, G.J.; Den Dunnen, J.T. Phage display screening without repetitious selection rounds. Anal. Biochem. 2012, 421, 622–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramaraju, H.; Miller, S.J.; Kohn, D.H. Dual-functioning peptides discovered by phage display increase the magnitude and specificity of BMSC attachment to mineralized biomaterials. Biomaterials 2017, 134, 1–12. [Google Scholar] [CrossRef]
- Ramaraju, H.; Kohn, D.H. Cell and material-specific phage display peptides increase IPS-MSC mediated bone and vasculature formation in vivo. Adv. Healthc. Mater. 2019, 8. [Google Scholar] [CrossRef]
- Shao, Z.; Zhang, X.; Pi, Y.; Wang, X.; Jia, Z.; Zhu, J.; Dai, L.; Chen, W.; Yin, L.; Chen, H.; et al. Polycaprolactone electrospun mesh conjugated with an MSC affinity peptide for MSC homing in vivo. Biomaterials 2012, 33, 3375–3387. [Google Scholar] [CrossRef]
- Meng, Q.; Man, Z.; Dai, L.; Huang, H.; Zhang, X.; Hu, X.; Shao, Z.; Zhu, J.; Zhang, J.; Fu, X.; et al. A composite scaffold of MSC affinity peptide-modified demineralized bone matrix particles and chitosan hydrogel for cartilage regeneration. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Man, Z.; Zhang, N.; Xin, H.; Li, Y.; Sun, T.; Sun, S. Biopanning of mouse bone marrow mesenchymal stem cell affinity for cyclic peptides. Mol. Med. Rep. 2019, 19, 407–413. [Google Scholar] [CrossRef]
- Teesalu, T.; Sugahara, K.N.; Ruoslahti, E. Mapping of vascular ZIP codes by phage display. In Methods in Enzymology; Academic Press Inc.: Orlando, FL, USA, 2012; Volume 503, pp. 35–56. [Google Scholar] [CrossRef]
- Soendergaard, M.; Newton-Northup, J.R.; Deutscher, S.L. In vivo phage display selection of an ovarian cancer targeting peptide for SPECT/CT imaging. Am. J. Nucl. Med. Mol. Imaging 2014, 4, 561–570. [Google Scholar]
- Hussain, S.; Joo, J.; Kang, J.; Kim, B.; Braun, G.B.; She, Z.G.; Kim, D.; Mann, A.P.; Mölder, T.; Teesalu, T.; et al. Antibiotic-loaded nanoparticles targeted to the site of infection enhance antibacterial efficacy. Nat. Biomed. Eng. 2018, 2, 95–103. [Google Scholar] [CrossRef]
- Meyers, S.R.; Hamilton, P.T.; Walsh, E.B.; Kenan, D.J.; Grinstaff, M.W. Endothelialization of titanium surfaces. Adv. Mater. 2007, 19, 2492–2498. [Google Scholar] [CrossRef]
- Naik, R.R.; Stringer, S.J.; Agarwal, G.; Jones, S.E.; Stone, M.O. Biomimetic synthesis and patterning of silver nanoparticles. Nat. Mater. 2002, 1, 169–172. [Google Scholar] [CrossRef]
- Wei, Z.; Maeda, Y.; Kanetsuki, Y.; Shi, M.; Matsui, H. Screening of oligopeptides that recognize inorganic crystalline facets of metal nanoparticles. Isr. J. Chem. 2019, 55, 749–755. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Li, Y.; Ruan, L.; Ye, X.; Murray, C.B.; Huang, Y. Platinum nanocrystals selectively shaped using facet-specific peptide sequences. Nat. Chem. 2011, 3, 393–399. [Google Scholar] [CrossRef]
- Kase, D.; Kulp, J.L.; Yudasaka, M.; Evans, J.S.; Iijima, S.; Shiba, K. Affinity selection of peptide phage libraries against single-wall carbon nanohorns identifies a peptide aptamer with conformational variability. Langmuir 2004, 20, 8939–8941. [Google Scholar] [CrossRef]
- Estephan, E.; Saab, M.-B.; Agarwal, V.; Cuisinier, F.J.G.; Larroque, C.; Gergely, C. Peptides for the biofunctionalization of silicon for use in optical sensing with porous silicon microcavities. Adv. Funct. Mater. 2011, 21, 2003–2011. [Google Scholar] [CrossRef]
- Luong, L.N.; Hong, S.I.; Patel, R.J.; Outslay, M.E.; Kohn, D.H. Spatial control of protein within biomimetically nucleated mineral. Biomaterials 2006, 27, 1175–1186. [Google Scholar] [CrossRef]
- Segvich, S.J.; Smith, H.C.; Kohn, D.H. The adsorption of preferential binding peptides to apatite-based materials. Biomaterials 2009, 30, 1287–1298. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Grainger, D.W. Drug/Device combinations for local drug therapies and infection prophylaxis. Biomaterials 2006, 27, 2450–2467. [Google Scholar] [CrossRef]
- Khoo, X.; O’Toole, G.A.; Nair, S.A.; Snyder, B.D.; Kenan, D.J.; Grinstaff, M.W. Staphylococcus aureus resistance on titanium coated with multivalent PEGylated-peptides. Biomaterials 2010, 31, 9285–9292. [Google Scholar] [CrossRef] [Green Version]
- Sanghvi, A.B.; Miller, K.P.H.; Belcher, A.M.; Schmidt, C.E. Biomaterials functionalization using a novel peptide that selectively binds to a conducting polymer. Nat. Mater. 2005, 4, 496–502. [Google Scholar] [CrossRef]
- Meyers, S.R.; Kenan, D.J.; Khoo, X.; Grinstaff, M.W. A bioactive stent surface coating that promotes endothelialization while preventing platelet adhesion. Biomacromolecules 2011, 12, 533–539. [Google Scholar] [CrossRef] [Green Version]
- Ramaraju, H.; Miller, S.J.; Kohn, D.H. Dual-functioning phage-derived peptides encourage human bone marrow cell-specific attachment to mineralized biomaterials. Connect. Tissue Res. 2014, 55, 160–163. [Google Scholar] [CrossRef] [Green Version]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davidson, T.A.; McGoldrick, S.J.; Kohn, D.H. Phage Display to Augment Biomaterial Function. Int. J. Mol. Sci. 2020, 21, 5994. https://doi.org/10.3390/ijms21175994
Davidson TA, McGoldrick SJ, Kohn DH. Phage Display to Augment Biomaterial Function. International Journal of Molecular Sciences. 2020; 21(17):5994. https://doi.org/10.3390/ijms21175994
Chicago/Turabian StyleDavidson, Thomas A., Samantha J. McGoldrick, and David H. Kohn. 2020. "Phage Display to Augment Biomaterial Function" International Journal of Molecular Sciences 21, no. 17: 5994. https://doi.org/10.3390/ijms21175994