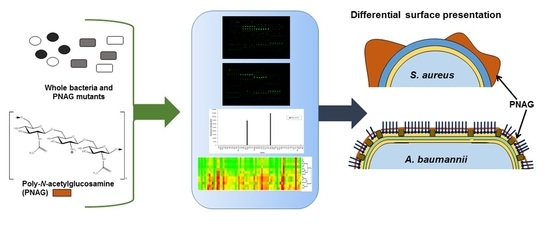
Glycomics Microarrays Reveal Differential In Situ Presentation of the Biofilm Polysaccharide Poly-N-acetylglucosamine on Acinetobacter baumannii and Staphylococcus aureus Cell Surfaces
Abstract
:1. Introduction
2. Results
2.1. Bacterial Strain Selections and Verification of Biofilm Production
2.2. Lectin Recognition of PNAG and Carbohydrate-mediated Binding Inhibition
2.3. PNAG Presentation and Accessibility In Situ on Bacterial Cell Surface
2.4. Carbohydrate Specificity of A. baumannii and S. aureus Strains
2.5. Bacterial Lectin Function in A. baumannii Biofilm Assembly
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Bacterial Strains and Culture
4.3. Biofilm Assays
4.4. Fluorescent Labeling of Bacteria
4.5. Assay for PNAG on Bacterial Cell Surface
4.6. Purification and Characterization of PNAG from S. aureus Mn8m
4.7. Fluorescent Labeling of PNAG
4.8. Lectin and Carbohydrate Microarray Construction
4.9. Microarray Incubation and Scanning
4.10. Microarray Data Extraction and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PNAG | poly-N-acetylglucosamine |
| MSSA | methicillin-sensitive S. aureus |
| MRSA | methicillin-resistant S. aureus |
| WGA | Wheatgerm agglutinin |
| sWGA | Succinylated WGA |
| eDNA | extracellular DNA |
| mAb | Monoclonal antibody |
| WT | Wild type |
| GlcNAc | N-acetylglucosamine |
| LTA | Lipoteichoic acid |
| Man | Mannose |
| IC50 | half maximal inhibitory concentration |
| HPA | Helix pomatia agglutinin |
| GSL-II | Griffonia simplicifolia lectin-II |
| DSA | Datura stramonium agglutinin |
| AAL | Aleuria aurantia lectin |
| MPA | Maclura pomifera agglutinin |
| SSEA3 | stage specific embryonic antigen 3 |
| DBA | Dolichos biflorus agglutinin |
| ACA | Amaranthus caudatus agglutinin |
| AMA | Arum maculatum agglutinin |
| LacNAc | N-acetyllactosamine |
| CPA | Cicer arietinum agglutinin |
| VRA | Vigna radiata agglutinin |
| LPS | lipopolysaccharide |
| CPS | Capsular polysaccharide |
| 3SLac | 3′sialyllactose |
| 6SLac | 6′sialyllactose |
| LNT | lacto-N-tetraose |
| FnBP | fibronectin-binding protein |
| Lex | Lewis x |
| BGA | Blood group A |
| BGB | Blood group B |
| BHI | Brain Heart Infusion |
| HRP | Horseradish peroxidase |
References
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.-F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organisation. Report on the Burden of Endemic Health Care-Associated Infection Worldwide; World Health Organisation: Geneva, Switzerland, 2011. [Google Scholar]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPEpathogens. BioMed. Res. Int. 2016, 2016, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, M.H.; Shu, J.C.; Lin, L.P.; Chong, K.y.; Cheng, Y.W.; Du, J.F.; Liu, S.-T. Elucidating the crucial role of poly N-acetylglucosamine from Staphylococcus aureus in cellular adhesion and pathogenesis. PLoS ONE 2015, 10, e0124216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maira-Litrán, T.; Kropec, A.; Goldmann, D.; Pier, G.B. Biologic properties and vaccine potential of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Vaccine 2004, 22, 872–879. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Huang, T.-W.; Yang, Y.-S.; Kuo, S.-C.; Chen, C.-T.; Liu, C.-P.; Liu, Y.-M.; Chen, T.-L.; Chang, F.-Y.; Wu, S.-H.; et al. Biofilm formation is not associated with worse outcome in Acinetobacter baumannii bacteraemic pneumonia. Sci. Rep. 2018, 8, 7289. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Smyth, D.; Humphreys, H.; Robinson, D.A.; O’Gara, J.P. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 2007, 45, 1379–1388. [Google Scholar] [CrossRef] [Green Version]
- Pozzi, C.; Waters, E.M.; Rudkin, J.K.; Schaeffer, C.R.; Lohan, A.J.; Tong, P.; Loftus, B.J.; Pier, G.B.; Fey, P.D.; Massey, R.C.; et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012, 8, e1002626. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; DeLeo, F.R.; Otto, M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [CrossRef] [Green Version]
- Cerca, N.; Jefferson, K.K. Effect of growth conditions on poly-N-acetylglucosamine expression and biofilm formation in Escherichia coli. FEMS Microbiol. Lett. 2008, 283, 36–41. [Google Scholar] [CrossRef]
- Conlon, K.M.; Humphreys, H.; O’Gara, J.P. Icar encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 2002, 184, 4400–4408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, F. Staphylococcus and biofilms. Mol. Microbiol. 2002, 43, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, A.; Alvarez, O.; Alonsa, B.; Llovo, J. Lectin typing of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 1999, 48, 495–499. [Google Scholar] [CrossRef]
- Neu, R.T.; Kuhlicke, U. Fluorescence lectin bar-coding of glycoconjugates in the extracellular matrix of biofilm and bioaggregate forming microorganisms. Microorganisms 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.M.; Ernst, O.; Manes, N.P.; Oyler, B.L.; Fraser, I.D.C.; Goodlett, D.R.; Nita-Lazar, A. Multi-omics strategies uncover host–pathogen interactions. ACS Inf. Dis. 2019, 5, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.N.C.; Yildiz, F.H. Biofilm matrix proteins. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Passos da Silva, D.; Matwichuk, M.L.; Townsend, D.O.; Reichhardt, C.; Lamba, D.; Wozniak, D.J.; Parsek, M.R. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat. Commun. 2019, 10, 2183. [Google Scholar] [CrossRef]
- Hsu, K.-L.; Pilobello, K.T.; Mahal, L.K. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat. Chem. Biol. 2006, 2, 153–157. [Google Scholar] [CrossRef]
- Kilcoyne, M.; Twomey, M.E.; Gerlach, J.Q.; Kane, M.; Moran, A.P.; Joshi, L. Campylobacter jejuni strain discrimination and temperature-dependent glycome expression profiling by lectin microarray. Carbohydr. Res. 2014, 389, 123–133. [Google Scholar] [CrossRef]
- Yasuda, E.; Tateno, H.; Hirabarashi, J.; Iino, T.; Sako, T. Lectin microarray reveals binding profiles of lactobacillus casei strains in a comprehensive analysis of bacterial cell wall polysaccharides. Appl. Environ. Microbiol. 2011, 77, 4539–4546. [Google Scholar] [CrossRef] [Green Version]
- Flannery, A.; Gerlach, J.Q.; Joshi, L.; Kilcoyne, M. Assessing bacterial interactions using carbohydrate-based microarrays. Microarrays 2015, 4, 690–713. [Google Scholar] [CrossRef] [PubMed]
- Horsburgh, M.J.; Aish, J.L.; White, I.J.; Shaw, L.; Lithgow, J.K.; Foster, S.J. Σb modulates virulence determinant expression and stress resistance: Characterization of a functional rsbu strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 2002, 184, 5457–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenney, D.; Pouliot, K.L.; Wang, Y.; Murthy, V.; Ulrich, M.; Döring, G.; Lee, J.C.; Goldmann, D.A.; Pier, G.B. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 1999, 284, 1523–1527. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, K.K.; Cramton, S.E.; Götz, F.; Pier, G.B. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of icar. Mol. Microbiol. 2003, 48, 889–899. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, F.; Humphreys, H.; O’Gara, J.P. Evidence for icaadbc-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 2005, 43, 1973–1976. [Google Scholar] [CrossRef] [Green Version]
- Houston, P.; Rowe, S.E.; Pozzi, C.; Waters, E.M.; O’Gara, J.P. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 2011, 79, 1153–1165. [Google Scholar] [CrossRef] [Green Version]
- Choi, A.H.K.; Slamti, L.; Avci, F.Y.; Pier, G.B.; Maira-Litrán, T. The pgaabcd locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 2009, 191, 5953–5963. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, C.A.; O’ Gara, J.P. Contribution of culture media and chemical properties of polystyrene tissue culture plates to biofilm development by Staphylococcus aureus. J. Med. Microbiol. 2004, 53, 1171–1173. [Google Scholar] [CrossRef]
- Lim, Y.; Jana, M.; Luong, T.T.; Lee, C.Y. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 2004, 186, 722–729. [Google Scholar] [CrossRef] [Green Version]
- Jefferson, K.K.; Pier, D.B.; Goldmann, D.A.; Pier, G.B. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 2004, 186, 2449–2456. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, G.B.; Marmont, L.S.; Howell, P.L. Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front. Microbiol. 2015, 6, 471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formosa-Dague, C.; Feuillie, C.; Beaussart, A.; Derclaye, S.; Kucharíková, S.; Lasa, I.; Van Dijck, P.; Dufrêne, Y.F. Sticky matrix: Adhesion mechanism of the staphylococcal polysaccharide intercellular adhesin. ACS Nano 2016, 10, 3443–3452. [Google Scholar] [CrossRef]
- Sanford, B.A.; Thomas, V.L.; Mattingly, S.J.; Ramsay, M.A.; Miller, M.M. Lectin-biotin assay for slime present in in situ biofilm produced by Staphylococcus epidermidis using transmission electron microscopy (TEM). J. Ind. Microbiol. 1995, 15, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Begun, J.; Gaiani, J.M.; Rohde, H.; Mack, D.; Calderwood, S.B.; Ausubel, F.M.; Sifri, C.D. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007, 3, e57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, Y.; Rocha, J.; Hael, A.L.; Van Gestel, J.; Vlamakis, H.; Cywes-Bentley, C.; Cubillos-Ruiz, J.R.; Pier, G.B.; Gilmore, M.S.; Kolter, R.; et al. PolyGlcNAc-containing exopolymers enable surface penetration by non-motile Enterococcus faecalis. PLoS Pathog. 2019, 15, e1007571. [Google Scholar] [CrossRef] [Green Version]
- Cerca, N.; Jefferson, K.K.; Oliveira, R.; Pier, G.B.; Azeredo, J. Comparative antibody-mediated phagocytosis of Staphylococcus epidermidis cells grown in a biofilm or in the planktonic state. Infect. Immun. 2006, 74, 4849–4855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sizemore, R.K.; Caldwell, J.J.; Kendrick, A.S. Alternate Gram staining technique using a fluorescent lectin. Appl. Environ. Microbiol. 1990, 56, 2245–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maira-Litrán, T.; Kropec, A.; Abeygunawardana, C.; Joyce, J.; Mark, G.; Goldmann, D.A.; Pier, G.B. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 2002, 70, 4433–4440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginsburg, I. Role of lipoteichoic acid in infection and inflammation. Lancet Infect. Dis. 2002, 2, 171–179. [Google Scholar] [CrossRef]
- Duckworth, M.; Archibald, A.R.; Baddiley, J. Lipoteichoic acid and lipoteichoic acid carrier in Staphylococcus aureus H. FEBS Lett. 1975, 53, 176–179. [Google Scholar] [CrossRef] [Green Version]
- Vinogradov, E.; Sadovskaya, I.; Li, J.; Jabbouri, S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus aureus Mn8m, a biofilm forming strain. Carbohydr. Res. 2006, 341, 738–743. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, Fnbpa and Fnbpb. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef] [Green Version]
- Bæk, K.T.; Gründling, A.; Mogensen, R.G.; Thøgersen, L.; Petersen, A.; Paulander, W.; Frees, D. Β-lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the clpxp protease. Antimicrob. Agents Chemother. 2014, 58, 4593–4603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, M.; Yamada, S.; Ishidoshiro, A.; Oyamada, Y.; Ito, H.; Yamagishi, J.-I. Cell-wall thickness: Possible mechanism of acriflavine resistance in meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 2009, 58, 331–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.-H.; Wang, S.-K.; Lin, C.-W.; Wang, C.-C.; Wong, C.-H.; Wu, C.-Y. Effects of neighboring glycans on antibody-carbohydrate interaction. Angew. Chem. Int. Ed. 2011, 50, 1608–1612. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Arbatsky, N.P.; Shneider, M.M.; Kenyon, J.J.; Shashkov, A.S.; Popova, A.V.; Miroshnikov, K.A.; Volozhantsev, N.V.; Knirel, Y.A. Structure of the neutral capsular polysaccharide of Acinetobacter baumannii NIPH146 that carries the kl37 capsule gene cluster. Carbohydr. Res. 2015, 413, 12–15. [Google Scholar] [CrossRef]
- MacLean, L.L.; Perry, M.B.; Chen, W.; Vinogradov, E. The structure of the polysaccharide O-chain of the LPS from Acinetobacter baumannii strain ATCC 17961. Carbohydr. Res. 2009, 344, 474–478. [Google Scholar] [CrossRef]
- Utratna, M.; Annuk, H.; Gerlach, J.Q.; Lee, Y.C.; Kane, M.; Kilcoyne, M.; Joshi, L. Rapid screening for specific glycosylation and pathogen interactions on a 78 species avian egg white glycoprotein microarray. Sci. Rep. 2017, 7, 6477. [Google Scholar] [CrossRef]
- Iyer, R.N.; Goldstein, I.J. Quantitative studies on the interaction of Concanavalin A, the carbohydrate-binding protein of the jack bean, with model carbohydrate-protein conjugates. Immunochemistry 1973, 10, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Oyelaran, O.; Li, Q.; Farnsworth, D.; Gildersleeve, J.C. Microarrays with varying carbohydrate density reveal distinct subpopulations of serum antibodies. J. Proteome Res. 2009, 8, 3529–3538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wacklin, P.; Mäkivuokko, H.; Alakulppi, N.; Nikkilä, J.; Tenkanen, H.; Räbinä, J.; Partanen, J.; Aranko, K.; Mättö, J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE 2011, 6, e20113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurjadi, D.; Lependu, J.; Kremsner, P.G.; Zanger, P. Staphylococcus aureus throat carriage is associated with ABO-/secretor status. J. Infect. 2012, 65, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Saadi, A.T.; Weir, D.M.; Poxton, I.R.; Stewart, J.; Essery, S.D.; Caroline Blackwell, C.; Raza, M.W.; Busuttil, A. Isolation of an adhesin from Staphylococcus aureus that binds Lewis blood group antigen and its relevance to sudden infant death syndrome. FEMS Immunol. Med. Microbiol. 1994, 8, 315–320. [Google Scholar] [CrossRef]
- Piroth, L.; Que, Y.-A.; Widmer, E.; Panchaud, A.; Piu, S.; Entenza, J.M.; Moreillon, P. The fibrinogen- and fibronectin-binding domains of Staphylococcus aureus fibronectin-binding protein a synergistically promote endothelial invasion and experimental endocarditis. Infect. Immun. 2008, 76, 3824–3831. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.S.; Kim, B.S.; Park, H.; Park, H.-Y.; Choi, H.-D.; Kim, H.H. Type and branched pattern of N-glycans and their structural effect on the chicken egg allergen ovotransferrin: A comparison with ovomucoid. Glycoconjug. J. 2014, 31, 41–50. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Koeleman, C.A.M.; Uh, H.-W.; Stam, J.C.; Van Heemst, D.; Maier, A.B.; Houwing-Duistermaat, J.J.; Hensbergen, P.J.; Slagboom, P.E.; Deelder, A.M.; et al. Targeted biomarker discovery by high throughput glycosylation profiling of human plasma alpha1-antitrypsin and immunoglobulin A. PLoS ONE 2013, 8, e73082. [Google Scholar] [CrossRef] [Green Version]
- Dengler, V.; Foulston, L.; DeFrancesco, A.S.; Losick, R. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2015, 197, 3779–3787. [Google Scholar] [CrossRef] [Green Version]
- Gottenbos, B.; Grijpma, D.W.; Van der Mei, H.C.; Feijen, J.; Busscher, H.J. Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2001, 48, 7–13. [Google Scholar] [CrossRef]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef] [Green Version]
- Cabral, M.P.; Soares, N.C.; Aranda, J.; Parreira, J.R.; Rumbo, C.; Poza, M.; Valle, J.; Calamia, V.; Lasa, Í.; Bou, G. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 2011, 10, 3399–3417. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Whitney, A.R.; DeLeo, F.R.; Otto, M. Polysaccharide intercellular adhesin (pia) protects staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004, 6, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Gaddy, J.A.; Actis, L.A. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 2009, 4, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Kilcoyne, M.; Gerlach, J.Q.; Kane, M.; Joshi, L. Surface chemistry and linker effects on lectin–carbohydrate recognition for glycan microarrays. Anal. Methods 2012, 4, 2721–2728. [Google Scholar] [CrossRef] [Green Version]
- Kelly-Quintos, C.; Cavacini, L.A.; Posner, M.R.; Goldmann, D.; Pier, G.B. Characterization of the opsonic and protective activity against Staphylococcus aureus of fully human monoclonal antibodies specific for the bacterial surface polysaccharide poly-N-acetylglucosamine. Infect. Immun. 2006, 74, 2742–2750. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Species and Strain | Sensitivity | Additive | Biofilm Effect | Major Biofilm Type | ||
|---|---|---|---|---|---|---|
| Protein | eDNA | PNAG | ||||
| S. aureus 8325-4 | MSSA | Glc | ↑ | ✓ | ||
| NaCl | ↑ | ✓ | ||||
| S. aureus Mn8m | MSSA | Glc | ↑ | ✓ | ||
| NaCl | n.d. | n.d. | n.d. | n.d. | ||
| S. aureus BH1CC | MRSA | Glc | ↑ | ✓ | ✓ | |
| NaCl | ↓ | n.d. | n.d. | n.d. | ||
| A. baumannii S1 | n.d. | Glc | n.d. | ✓ | ||
| NaCl | n.d. | n.d. | n.d. | n.d. | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flannery, A.; Le Berre, M.; Pier, G.B.; O’Gara, J.P.; Kilcoyne, M. Glycomics Microarrays Reveal Differential In Situ Presentation of the Biofilm Polysaccharide Poly-N-acetylglucosamine on Acinetobacter baumannii and Staphylococcus aureus Cell Surfaces. Int. J. Mol. Sci. 2020, 21, 2465. https://doi.org/10.3390/ijms21072465
Flannery A, Le Berre M, Pier GB, O’Gara JP, Kilcoyne M. Glycomics Microarrays Reveal Differential In Situ Presentation of the Biofilm Polysaccharide Poly-N-acetylglucosamine on Acinetobacter baumannii and Staphylococcus aureus Cell Surfaces. International Journal of Molecular Sciences. 2020; 21(7):2465. https://doi.org/10.3390/ijms21072465
Chicago/Turabian StyleFlannery, Andrea, Marie Le Berre, Gerald B. Pier, James P. O’Gara, and Michelle Kilcoyne. 2020. "Glycomics Microarrays Reveal Differential In Situ Presentation of the Biofilm Polysaccharide Poly-N-acetylglucosamine on Acinetobacter baumannii and Staphylococcus aureus Cell Surfaces" International Journal of Molecular Sciences 21, no. 7: 2465. https://doi.org/10.3390/ijms21072465