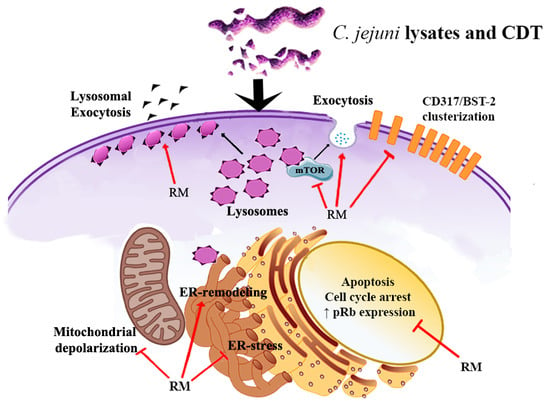
Rapamycin Re-Directs Lysosome Network, Stimulates ER-Remodeling, Involving Membrane CD317 and Affecting Exocytosis, in Campylobacter Jejuni-Lysate-Infected U937 Cells
Abstract
:1. Introduction
2. Results
2.1. Morphological Features, Cell Death, Absolute Count, and Evaluation of Cellular Division
2.2. ER Stress Evaluation
2.3. Prb Detection
2.4. Rapamycin Inhibition of MTORC1 Signalling Reduces CDT-Induced Distension, Cell Death and Proliferation
2.5. Rapamycin Stimulates ER-Remodeling, Increases Lysosome Number, Modifies Their Distribution, Decreasing Prb and Bcl-2 Content
2.6. Evaluation of Membrane Microdomains
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Growth Conditions of Bacterial Strains and Cell Lysate Preparation
4.3. Pre-Treatment of U937 Cells with Rapamycin
4.4. Pre-Treatment of U937 Cells With C. jejuni Lysates
4.5. Detection of Cytotoxin Activity in C. jejuni Lysates
4.6. Morphological Feature Evaluation
4.7. Flow Cytometry (FCM) and Confocal Microscopy (CM) Stainings
4.7.1. Flow Cytometric Detection of Cell Death and Flow Cytometric Absolute Count
4.7.2. Evaluation of Cellular Division by CFSE Staining
4.7.3. Assessment of Lysosomal Involvement
4.7.4. Determination of Mitochondria and Mitochondrial Membrane Potential (∆Ψm)
4.7.5. ER Stress Evaluation
4.7.6. CD317 Expression Evaluation
4.7.7. Intracellular Detection of Prb and Bcl-2 Antigens
4.7.8. Extracellular Vesicle Detection
4.7.9. Cytometric Investigations
4.7.10. Confocal Microscopy Analyses
4.8. RNA Isolation and cDNA Synthesis
4.9. Quantitative Real-Time PCR
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| C. jejuni | Campylobacter jejuni |
| AnxV | Annexin V |
| Atg | Autophagy-Related Proteins |
| CDT | Cytolethal Distending Toxin |
| CFSE | Carboxyfluoresceinsuccinimidyl ester |
| ER | Endoplasmic Reticulum |
| EVs | Extracellular vesicles |
| GBS | Guillain-Barré syndrome |
| LTDR | LysoTracker Deep Red |
| MFI | Mean Fluorescence Intensity |
| MVB | Multivesicular body |
| pRb | Retinoblastoma Protein |
| ReA | reactive arthritis |
| RM | Rapamycin |
| TGN | Trans-Golgi Network |
| TMRE | Tetramethylrhodamine ethyl ester perchlorate |
| UPR | Unfolded protein response |
References
- Shen, H.M.; Mizushima, N. At the end of the autophagic road: An emerging understanding of lysosomal functions in autophagy. Trends Biochem. Sci. 2014, 39, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Py, B.F.; Lipinski, M.M.; Yuan, J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy 2007, 3, 117–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Green, D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef] [PubMed]
- Lum, J.J.; Bauer, D.E.; Kong, M.; Harris, M.H.; Li, C.; Lindsten, T.; Thompson, C.B. Growth Factor Regulation of Autophagy and Cell Survival in the Absence of Apoptosis. Cell 2005, 120, 237–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, S.; Kanaseki, T.; Mizushima, N.; Mizuta, T.; Arakawa-Kobayashi, S.; Thompson, C.B.; Tsujimoto, Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004, 6, 1221–1228. [Google Scholar] [CrossRef]
- Booth, L.A.; Roberts, J.L.; Dent, P. The role of cell signaling in the crosstalk between autophagy and apoptosis in the regulation of tumor cell survival in response to sorafenib and neratinib. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef]
- Nikoletopoulou, V.; Markaki, M.; Palikaras, K.; Tavernarakis, N. Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta - Mol. Cell Res. 2013, 1833, 3448–3459. [Google Scholar] [CrossRef] [Green Version]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef]
- Sehgal, S.N. Sirolimus: Its discovery, biological properties, and mechanism of action. Transplant. Proc. 2003, 35, 7S–14S. [Google Scholar] [CrossRef]
- Babcock, J.T.; Nguyen, H.B.; He, Y.; Hendricks, J.W.; Wek, R.C.; Quilliam, L.A. Mammalian Target of Rapamycin Complex 1 (mTORC1) enhances bortezomib-induced death in Tuberous Sclerosis Complex (TSC)-null cells by a c-MYC-dependent induction of the unfolded protein response. J. Biol. Chem. 2013, 288, 15687–15698. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Nakajima, S.; Saito, Y.; Takahashi, S.; Katoh, R.; Kitamura, M. MTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1-JNK pathway. Cell Death Differ. 2012, 19, 310–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Y.; Luo, X.; Yang, Y.; Dai, Z.; Wu, G.; Wu, Z. Endoplasmic reticulum stress-induced apoptosis in intestinal epithelial cells: A feed-back regulation by mechanistic target of rapamycin complex 1 (mTORC1). J. Anim. Sci. Biotechnol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Boesze-Battaglia, K.; Alexander, D.; Dlakić, M.; Shenker, B.J. A Journey of Cytolethal Distending Toxins through Cell Membranes. Front. Cell. Infect. Microbiol. 2016, 6, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, K.N.; Kober, D.L.; Su, A.; Brett, T.J. Limiting Respiratory Viral Infection by Targeting Antiviral and Immunological Functions of BST-2/Tetherin: Knowledge and Gaps. BioEssays 2018, 40, 1800086. [Google Scholar] [CrossRef] [PubMed]
- Kupzig, S.; Korolchuk, V.; Rollason, R.; Sugden, A.; Wilde, A.; Banting, G. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 2003, 4, 694–709. [Google Scholar] [CrossRef]
- Masuyama, N.; Kuronita, T.; Tanaka, R.; Muto, T.; Hirota, Y.; Takigawa, A.; Fujita, H.; Aso, Y.; Amano, J.; Tanaka, Y. HM1.24 Is Internalized from Lipid Rafts by Clathrin-mediated Endocytosis through Interaction with α-Adaptin. J. Biol. Chem. 2009, 284, 15927–15941. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Li, X.; Liu, L.; Li, J.; Chen, Q.; Huang, S.; Li, Y.; Wan, X. Vaccination with a DNA vaccine encoding CD317-targeting HBs antigen elicits enhanced immunity in mice. Biochem. Biophys. Res. Commun. 2018, 504, 865–870. [Google Scholar] [CrossRef]
- Yi, E.; Oh, J.; Kang, H.R.; Song, M.J.; Park, S.H. BST2 inhibits infection of influenza A virus by promoting apoptosis of infected cells. Biochem. Biophys. Res. Commun. 2019. [Google Scholar] [CrossRef]
- Neil, S.J.D.; Zang, T.; Bieniasz, P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Edgar, J.R.; Manna, P.T.; Nishimura, S.; Banting, G.; Robinson, M.S. Tetherin is an exosomal tether. Elife 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Mead, P.S.; Slutsker, L.; Dietz, V.; McCaig, L.F.; Bresee, J.S.; Shapiro, C.; Griffin, P.M.; Tauxe, R. V Food-related illness and death in the United States. Emerg. Infect. Dis. 1999, 5, 607–625. [Google Scholar] [CrossRef] [PubMed]
- Corry, J.E.L.; Atabay, H.I. Poultry as a source of Campylobacter and related organisms. J. Appl. Microbiol. 2001, 90, 96S–114S. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.M.; Lior, H. A new heat-labile cytolethal distending toxin (CLDT) produced by Campylobacter spp. Microb. Pathog. 1988, 4, 115–126. [Google Scholar] [CrossRef]
- Lin, C.-D.; Lai, C.-K.; Lin, Y.-H.; Hsieh, J.-T.; Sing, Y.-T.; Chang, Y.-C.; Chen, K.-C.; Wang, W.-C.; Su, H.-L.; Lai, C.-H. Cholesterol Depletion Reduces Entry of Campylobacter jejuni Cytolethal Distending Toxin and Attenuates Intoxication of Host Cells. Infect. Immun. 2011, 79, 3563–3575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohara, M.; Oswald, E.; Sugai, M. Cytolethal Distending Toxin: A Bacterial Bullet Targeted to Nucleus. J. Biochem. 2004, 136, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Cope, L.D.; Lumbley, S.; Latimer, J.L.; Klesney-Tait, J.; Stevens, M.K.; Johnson, L.S.; Purven, M.; Munson, R.S.; Lagergard, T.; Radolf, J.D.; et al. A diffusible cytotoxin of Haemophilus ducreyi. Proc. Natl. Acad. Sci. USA 1997, 94, 4056–4061. [Google Scholar] [CrossRef] [Green Version]
- Young, V.B.; Chien, C.; Knox, K.A.; Taylor, N.S.; Schauer, D.B.; Fox, J.G. Cytolethal Distending Toxin in Avian and Human Isolates of Helicobacter pullorum. J. Infect. Dis. 2000, 182, 620–623. [Google Scholar] [CrossRef] [Green Version]
- Okuda, J.; Kurazono, H.; Takeda, Y. Distribution of the cytolethal distending toxin A gene (cdtA) among species of Shigella and Vibrio, and cloning and sequencing of the cdt gene from Shigella dysenteriae. Microb. Pathog. 1995, 18, 167–172. [Google Scholar] [CrossRef]
- Zhang, B.; He, Y.; Xu, C.; Xu, L.; Feng, S.; Liao, M.; Ren, T. Cytolethal distending toxin (CDT) of the Haemophilus parasuis SC096 strain contributes to serum resistance and adherence to and invasion of PK-15 and PUVEC cells. Vet. Microbiol. 2012, 157, 237–242. [Google Scholar] [CrossRef]
- Whitehouse, C.A.; Balbo, P.B.; Pesci, E.C.; Cottle, D.L.; Mirabito, P.M.; Pickett, C.L. Campylobacter jejuni Cytolethal Distending Toxin Causes a G 2 -Phase Cell Cycle Block. Infect. Immun. 1998, 66, 1934–1940. [Google Scholar] [CrossRef] [Green Version]
- Nesic, D.; Stebbins, C.E. Mechanisms of Assembly and Cellular Interactions for the Bacterial Genotoxin CDT. PLoS Pathog. 2005, 1, e28. [Google Scholar] [CrossRef] [PubMed]
- Lindmark, B.; Rompikuntal, P.; Vaitkevicius, K.; Song, T.; Mizunoe, Y.; Uhlin, B.; Guerry, P.; Wai, S. Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol. 2009, 9, 220. [Google Scholar] [CrossRef] [Green Version]
- Bezine, E.; Vignard, J.; Mirey, G.; Bezine, E.; Vignard, J.; Mirey, G. The Cytolethal Distending Toxin Effects on Mammalian Cells: A DNA Damage Perspective. Cells 2014, 3, 592–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boesze-Battaglia, K.; Besack, D.; McKay, T.; Zekavat, A.; Otis, L.; Jordan-Sciutto, K.; Shenker, B.J. Cholesterol-rich membrane microdomains mediate cell cycle arrest induced by Actinobacillus actinomycetemcomitans cytolethal-distending toxin. Cell. Microbiol. 2006, 8, 823–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boesze-Battaglia, K. Isolation of membrane rafts and signaling complexes. Methods Mol. Biol. 2006, 332, 169–179. [Google Scholar] [PubMed] [Green Version]
- Damek-Poprawa, M.; Jang, J.Y.; Volgina, A.; Korostoff, J.; DiRienzo, J.M. Localization of Aggregatibacter actinomycetemcomitans Cytolethal Distending Toxin Subunits during Intoxication of Live Cells. Infect. Immun. 2012, 80, 2761–2770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.W.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019, 68, 289–300. [Google Scholar] [CrossRef] [Green Version]
- Haghjoo, E.; Galán, J.E. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 4614–4619. [Google Scholar] [CrossRef] [Green Version]
- Nath, G.; Gulati, A.K.; Shukla, V.K. Role of bacteria in carcinogenesis, with special reference to carcinoma of the gallbladder. World J. Gastroenterol. 2010. [Google Scholar] [CrossRef]
- Shenker, B.J.; McKay, T.; Datar, S.; Miller, M.; Chowhan, R.; Demuth, D. Actinobacillus actinomycetemcomitans immunosuppressive protein is a member of the family of cytolethal distending toxins capable of causing a G2 arrest in human T cells. J. Immunol. 1999, 162, 4773–4780. [Google Scholar]
- Sandra Trott; Reinhard Bauer; Hans-Joachim Knackmuss; Andreas Stolz Campylobacter upsaliensis exerts a cytolethal distending toxin effect on HeLa cells and T lymphocytes. Microbiology 2001, 1815–1824.
- Kostia, S.; Veijalainen, P.; Hirvi, U.; Hänninen, M.L. Cytolethal distending toxin B gene (cdtB) homologues in taxa 2, 3 and 8 and in six canine isolates of Helicobacter sp. flexispira. J. Med. Microbiol. 2003, 52, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Nougayrède, J.P.; Taieb, F.; De Rycke, J.; Oswald, E. Cyclomodulins: Bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 2005, 13, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Biasoli, D.; Kahn, S.A.; Cornélio, T.A.; Furtado, M.; Campanati, L.; Chneiweiss, H.; Moura-Neto, V.; Borges, H.L. Retinoblastoma protein regulates the crosstalk between autophagy and apoptosis, and favors glioblastoma resistance to etoposide. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Canonico, B.; di Sario, G.; Cesarini, E.; Campana, R.; Luchetti, F.; Zamai, L.; Ortolani, C.; Nasoni, M.G.; Baffone, W.; Papa, S. Monocyte response to different Campylobacter jejuni lysates involves endoplasmic reticulum stress and the lysosomal–mitochondrial axis: When cell death is better than cell survival. Toxins (Basel) 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Wrigley, B.J.; Lip, G.Y.H.; Shantsila, E. The role of monocytes and inflammation in the pathophysiology of heart failure. Eur. J. Heart Fail. 2011, 13, 1161–1171. [Google Scholar] [CrossRef]
- Guerra, L.; Guidi, R.; Frisan, T. Do bacterial genotoxins contribute to chronic inflammation, genomic instability and tumor progression? FEBS J. 2011, 278, 4577–4588. [Google Scholar] [CrossRef] [Green Version]
- Eshraghi, A.; Maldonado-Arocho, F.J.; Gargi, A.; Cardwell, M.M.; Prouty, M.G.; Blanke, S.R.; Bradley, K.A. Cytolethal distending toxin family members are differentially affected by alterations in host glycans and membrane cholesterol. J. Biol. Chem. 2010, 285, 18199–18207. [Google Scholar] [CrossRef] [Green Version]
- Rabin, S.D.P.; Flitton, J.G.; Demuth, D.R. Aggregatibacter actinomycetemcomitans cytolethal distending toxin induces apoptosis in nonproliferating macrophages by a phosphatase-independent mechanism. Infect. Immun. 2009, 77, 3161–3169. [Google Scholar] [CrossRef] [Green Version]
- Choo, A.Y.; Yoon, S.O.; Sang, G.K.; Roux, P.P.; Blenis, J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci. USA 2008. [Google Scholar] [CrossRef] [Green Version]
- Komiya, T.; Memmott, R.M.; Blumenthal, G.M.; Bernstein, W.; Ballas, M.S.; De Chowdhury, R.; Chun, G.; Peer, C.J.; Figg, W.D.; Liewehr, D.J.; et al. A phase I/II study of pemetrexed with sirolimus in advanced, previously treated non-small cell lung cancer. Transl. Lung Cancer Res. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, P.; Wallin, P.; Sjöwall, C. Clinical experience of sirolimus regarding efficacy and safety in systemic lupus erythematosus. Front. Pharmacol. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohankumar, V.; Ramalingam, S.; Chidambaranathan, G.P.; Prajna, L. Autophagy induced by type III secretion system toxins enhances clearance of Pseudomonas aeruginosa from human corneal epithelial cells. Biochem. Biophys. Res. Commun. 2018. [Google Scholar] [CrossRef] [PubMed]
- Canonico, B.; Campana, R.; Luchetti, F.; Arcangeletti, M.; Betti, M.; Cesarini, E.; Ciacci, C.; Vittoria, E.; Galli, L.; Papa, S.; et al. Campylobacter jejuni cell lysates differently target mitochondria and lysosomes on HeLa cells. Apoptosis 2014, 19, 1225–1242. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Pua, H.H.; Li, Q.-J.; He, Y.-W. Autophagy Regulates Endoplasmic Reticulum Homeostasis and Calcium Mobilization in T Lymphocytes. J. Immunol. 2011, 186, 1564–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tentaku, A.; Shimohata, T.; Hatayama, S.; Kido, J.; Nguyen, A.Q.; Kanda, Y.; Fukushima, S.; Uebanso, T.; Iwata, T.; Mawatari, K.; et al. Host cellular unfolded protein response signaling regulates Campylobacter jejuni invasion. PLoS ONE 2018, 13, e0205865. [Google Scholar] [CrossRef] [PubMed]
- Nicolay, B.N.; Dyson, N.J. The multiple connections between pRB and cell metabolism. Curr. Opin. Cell Biol. 2013, 25, 735–740. [Google Scholar] [CrossRef] [Green Version]
- D’Angelo, B.; Astarita, C.; Boffo, S.; Massaro-Giordano, M.; Antonella Ianuzzi, C.; Caporaso, A.; Macaluso, M.; Giordano, A. LPS-induced inflammatory response triggers cell cycle reactivation in murine neuronal cells through retinoblastoma proteins induction. Cell Cycle 2017, 16, 2330–2336. [Google Scholar] [CrossRef] [Green Version]
- Flatt, P.M.; Tang, L.J.; Scatena, C.D.; Szak, S.T.; Pietenpol, J.A. p53 regulation of G(2) checkpoint is retinoblastoma protein dependent. Mol. Cell. Biol. 2000, 20, 4210–4223. [Google Scholar] [CrossRef] [Green Version]
- Fingar, D.C.; Blenis, J. Target of rapamycin (TOR): An integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 2004, 23, 3151–3171. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Syed, T.W.; Liu, R.; Yu, J. Role of Endoplasmic Reticulum Stress, Autophagy, and Inflammation in Cardiovascular Disease. Front. Cardiovasc. Med. 2017, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Sagua, R.; Parra, V.; Ortiz-Sandoval, C.; Navarro-Marquez, M.; Rodríguez, A.E.; Diaz-Valdivia, N.; Sanhueza, C.; Lopez-Crisosto, C.; Tahbaz, N.; Rothermel, B.A.; et al. Caveolin-1 impairs PKA-DRP1-mediated remodelling of ER–mitochondria communication during the early phase of ER stress. Cell Death Differ. 2019, 26, 1195–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korolchuk, V.I.; Rubinsztein, D.C. Regulation of autophagy by lysosomal positioning. Autophagy 2011, 7, 927–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billcliff, P.G.; Rollason, R.; Prior, I.; Owen, D.M.; Gaus, K.; Banting, G. CD317/tetherin is an organiser of membrane microdomains. J. Cell Sci. 2013, 126, 1553–1564. [Google Scholar] [CrossRef] [Green Version]
- Hogue, I.B.; Grover, J.R.; Soheilian, F.; Nagashima, K.; Ono, A. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 2011, 85, 9749–9766. [Google Scholar] [CrossRef] [Green Version]
- Mahauad-Fernandez, W.D.; Okeoma, C.M. The role of BST-2/Tetherin in host protection and disease manifestation. Immun. Inflamm. Dis. 2016, 4, 4–23. [Google Scholar] [CrossRef]
- Anikeeva, N.; Sykulev, Y. Mechanisms controlling granule-mediated cytolytic activity of cytotoxic T lymphocytes. Immunol. Res. 2011, 51, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Jongsma, M.L.L.M.; Berlin, I.; Wijdeven, R.H.H.M.; Janssen, L.; Janssen, G.M.M.C.; Garstka, M.A.A.; Janssen, H.; Mensink, M.; van Veelen, P.A.A.; Spaapen, R.M.M.; et al. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell 2016, 166, 152–166. [Google Scholar] [CrossRef] [Green Version]
- Reed, S.E.; Hodgson, L.R.; Song, S.; May, M.T.; Kelly, E.E.; McCaffrey, M.W.; Mastick, C.C.; Verkade, P.; Tavare, J.M. A role for Rab14 in the endocytic trafficking of GLUT4 in 3T3-L1 adipocytes. J. Cell Sci. 2013, 126, 1931–1941. [Google Scholar] [CrossRef] [Green Version]
- Rojo Pulido, I.; Nightingale, T.D.; Darchen, F.; Seabra, M.C.; Cutler, D.F.; Gerke, V. Myosin Va acts in concert with Rab27a and MyRIP to regulate acute von-Willebrand factor release from endothelial cells. Traffic 2011, 12, 1371–1382. [Google Scholar] [CrossRef]
- Sadacca, L.A.; Bruno, J.; Wen, J.; Xiong, W.; Mcgraw, T.E. Specialized sorting of GLUT4 and its recruitment to the cell surface are independently regulated by distinct Rabs. Mol. Biol. Cell 2013, 24, 2544–2557. [Google Scholar] [CrossRef] [PubMed]
- Wasmeier, C.; Hume, A.N.; Bolasco, G.; Seabra, M.C. Melanosomes at a glance. J. Cell Sci. 2008, 121, 3995–3999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henne, W.M. Organelle remodeling at membrane contact sites. J. Struct. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I. Open questions: Why should we care about ER-phagy and ER remodelling? BMC Biol. 2018, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Chiramel, A.I.; Dougherty, J.D.; Nair, V.; Robertson, S.J.; Best, S.M. FAM134B, the Selective Autophagy Receptor for Endoplasmic Reticulum Turnover, Inhibits Replication of Ebola Virus Strains Makona and Mayinga. J. Infect. Dis. 2016, 214, S319–S325. [Google Scholar] [CrossRef] [Green Version]
- Lennemann, N.J.; Rhein, B.A.; Ndungo, E.; Chandran, K.; Qiu, X.; Maury, W. Comprehensive functional analysis of N-linked glycans on ebola virus GP1. MBio 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Moretti, J.; Roy, S.; Bozec, D.; Martinez, J.; Chapman, J.R.; Ueberheide, B.; Lamming, D.W.; Chen, Z.J.; Horng, T.; Yeretssian, G.; et al. STING Senses Microbial Viability to Orchestrate Stress-Mediated Autophagy of the Endoplasmic Reticulum. Cell 2017, 171, 809–823.e13. [Google Scholar] [CrossRef]
- Loi, M.; Fregno, I.; Guerra, C.; Molinari, M. Eat it right: Er-phagy and recover-phagy. Biochem. Soc. Trans. 2018, 46, 699–706. [Google Scholar] [CrossRef] [Green Version]
- Helle, S.C.J.; Kanfer, G.; Kolar, K.; Lang, A.; Michel, A.H.; Kornmann, B. Organization and function of membrane contact sites. Biochim. Biophys. Acta - Mol. Cell Res. 2013. [Google Scholar] [CrossRef] [Green Version]
- Bravo-Sagua, R.; López-Crisosto, C.; Parra, V.; Rodriguez-Peña, M.; Rothermel, B.A.; Quest, A.F.G.; Lavandero, S. MTORC1 inhibitor rapamycin and ER stressor tunicamycin induce differential patterns of ER-mitochondria coupling. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Haupt, Y.; Rowan, S.; Oren, M. p53-mediated apoptosis in HeLa cells can be overcome by excess pRB. Oncogene 1995, 10, 1563–1571. [Google Scholar] [PubMed]
- Popowski, M.; Ferguson, H.A.; Sion, A.M.; Koller, E.; Knudsen, E.; Van Den Berg, C.L. Stress and IGF-I differentially control cell fate through mammalian target of rapamycin (mTOR) and retinoblastoma protein (pRB). J. Biol. Chem. 2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Zamudio, R.I.; Robinson, L.; Roux, P.F.; Bischof, O. SnapShot: Cellular Senescence Pathways. Cell 2017, 170, 816–816.e1. [Google Scholar] [CrossRef] [PubMed]
- Cabukusta, B.; Neefjes, J. Mechanisms of lysosomal positioning and movement. Traffic 2018, 19, 761–769. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Saiki, S.; Lichtenberg, M.; Siddiqi, F.H.; Roberts, E.A.; Imarisio, S.; Jahreiss, L.; Sarkar, S.; Futter, M.; Menzies, F.M.; et al. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 2011, 13, 453–462. [Google Scholar] [CrossRef]
- Raiborg, C.; Wenzel, E.M.; Pedersen, N.M.; Olsvik, H.; Schink, K.O.; Schultz, S.W.; Vietri, M.; Nisi, V.; Bucci, C.; Brech, A.; et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature 2015. [Google Scholar] [CrossRef]
- Rocha, N.; Kuijl, C.; Van Der Kant, R.; Janssen, L.; Houben, D.; Janssen, H.; Zwart, W.; Neefjes, J. Cholesterol sensor ORP1L organizes late endosomal contacts with the ER protein VAP that controls Rab7-RILP-p150 Glued and late endosomal positioning. J. Cell Biol. 2009. [Google Scholar] [CrossRef] [Green Version]
- Van Der Kant, R.; Neefjes, J. Small regulators, major consequences-Ca2+ and cholesterol at the endosome-ER interface. J. Cell Sci. 2014. [Google Scholar] [CrossRef] [Green Version]
- Rowland, A.A.; Chitwood, P.J.; Phillips, M.J.; Voeltz, G.K. ER contact sites define the position and timing of endosome fission. Cell 2014. [Google Scholar] [CrossRef] [Green Version]
- Rollason, R.; Dunstan, K.; Billcliff, P.G.; Bishop, P.; Gleeson, P.; Wise, H.; Digard, P.; Banting, G. Expression of HIV-1 Vpu Leads to Loss of the Viral Restriction Factor CD317/Tetherin from Lipid Rafts and Its Enhanced Lysosomal Degradation. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Mahauad-Fernandez, W.D.; Okeoma, C.M. BST-2: At the crossroads of viral pathogenesis and oncogenesis. Future Virol. 2016, 11, 127–140. [Google Scholar] [CrossRef]
- Inoki, K.; Li, Y.; Zhu, T.; Wu, J.; Guan, K.-L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002, 4, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR Signaling in Growth and Metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Threadgill, D.; Jobin, C. Campylobacter jejuni Induces Colitis Through Activation of Mammalian Target of Rapamycin Signaling. Gastroenterology 2012, 142, 86–95.e5. [Google Scholar] [CrossRef] [Green Version]
- Brando, B.; Barnett, D.; Janossy, G.; Mandy, F.; Autran, B.; Rothe, G.; Scarpati, B.; D’Avanzo, G.; D’Hautcourt, J.-L.; Lenkei, R.; et al. Cytofluorometric methods for assessing absolute numbers of cell subsets in blood. Cytometry 2000, 42, 327–346. [Google Scholar] [CrossRef]
- Chazotte, B. Labeling lysosomes in live cells with LysoTracker. Cold Spring Harb. Protoc. 2011, 2011, pdb.prot5571. [Google Scholar] [CrossRef]
- Canonico, B.; Cesarini, E.; Salucci, S.; Luchetti, F.; Falcieri, E.; Sario, G.D.; Palma, F.; Papa, S. Defective autophagy, mitochondrial clearance and lipophagy in niemann-pick type B lymphocytes. PLoS ONE 2016. [Google Scholar] [CrossRef]
- Luchetti, F.; Canonico, B.; Arcangeletti, M.; Guescini, M.; Cesarini, E.; Stocchi, V.; Degli Esposti, M.; Papa, S. Fas Signalling Promotes Intercellular Communication in T Cells. PLoS ONE 2012, 7, e35766. [Google Scholar] [CrossRef] [Green Version]
- Hogg, R.C.; Adams, D.J. An ATP-sensitive K(+) conductance in dissociated neurones from adult rat intracardiac ganglia. J. Physiol. 2001, 534, 713–720. [Google Scholar] [CrossRef]
- Ghasemi, M.; Khodaei, N.; Salari, S.; Eliassi, A.; Saghiri, R. Gating behavior of endoplasmic reticulum potassium channels of rat hepatocytes in diabetes. Iran. Biomed. J. 2014. [Google Scholar]
- Li, H.; Mao, G.; Carlson, J.; Leng, S.X. A novel flow cytometry-based tool for determining the efficiency of human cytomegalovirus infection in THP-1 derived macrophages. J. Virol. Methods 2015. [Google Scholar] [CrossRef] [PubMed]
- Brocco, D.; Lanuti, P.; Simeone, P.; Bologna, G.; Pieragostino, D.; Cufaro, M.C.; Graziano, V.; Peri, M.; Di Marino, P.; De Tursi, M.; et al. Circulating Cancer Stem Cell-Derived Extracellular Vesicles as a Novel Biomarker for Clinical Outcome Evaluation. J. Oncol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Diotallevi, A.; De Santi, M.; Ceccarelli, M.; Vitale, F.; Brandi, G.; Magnani, M. Leishmania infantum Induces Mild Unfolded Protein Response in Infected Macrophages. PLoS ONE 2016, 11, e0168339. [Google Scholar] [CrossRef] [PubMed]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canonico, B.; Cesarini, E.; Montanari, M.; Di Sario, G.; Campana, R.; Galluzzi, L.; Sola, F.; Gundogdu, O.; Luchetti, F.; Diotallevi, A.; et al. Rapamycin Re-Directs Lysosome Network, Stimulates ER-Remodeling, Involving Membrane CD317 and Affecting Exocytosis, in Campylobacter Jejuni-Lysate-Infected U937 Cells. Int. J. Mol. Sci. 2020, 21, 2207. https://doi.org/10.3390/ijms21062207
Canonico B, Cesarini E, Montanari M, Di Sario G, Campana R, Galluzzi L, Sola F, Gundogdu O, Luchetti F, Diotallevi A, et al. Rapamycin Re-Directs Lysosome Network, Stimulates ER-Remodeling, Involving Membrane CD317 and Affecting Exocytosis, in Campylobacter Jejuni-Lysate-Infected U937 Cells. International Journal of Molecular Sciences. 2020; 21(6):2207. https://doi.org/10.3390/ijms21062207
Chicago/Turabian StyleCanonico, Barbara, Erica Cesarini, Mariele Montanari, Gianna Di Sario, Raffaella Campana, Luca Galluzzi, Federica Sola, Ozan Gundogdu, Francesca Luchetti, Aurora Diotallevi, and et al. 2020. "Rapamycin Re-Directs Lysosome Network, Stimulates ER-Remodeling, Involving Membrane CD317 and Affecting Exocytosis, in Campylobacter Jejuni-Lysate-Infected U937 Cells" International Journal of Molecular Sciences 21, no. 6: 2207. https://doi.org/10.3390/ijms21062207