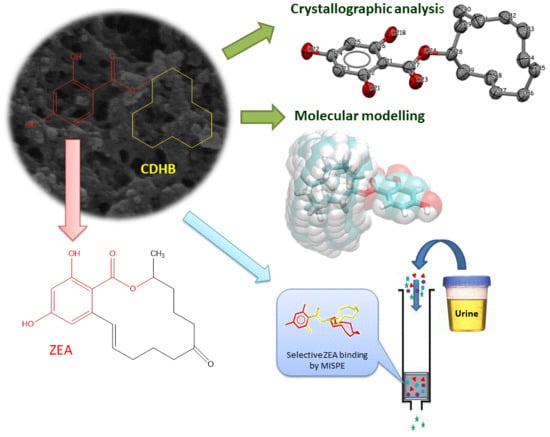
Towards A New Approach for the Description of Cyclo–2,4-Dihydroxybenzoate, A Substance Which Effectively Mimics Zearalenone in Imprinted Polymers Designed for Analyzing Selected Mycotoxins in Urine
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of CDHB
2.1.1. FT-IR Analysis
2.1.2. NMR
2.1.3. UV–VIS
2.1.4. Mass Spectrum
2.2. Characterization of Polymers
2.3. Quantum-Chemical Calculations
2.4. Crystal Structure of CDHB
2.5. Validation Method
Calibration Curves, Limits of Detection, and Quantification
2.6. Optimization of MISPE Parameters Influencing ZEA Adsorption
2.6.1. Sample pH
2.6.2. MIP Amount
2.6.3. Desorption
2.6.4. Kinetic Study
2.6.5. Adsorption Isotherm
2.6.6. Selectivity
2.7. Effectiveness of Isolating ZEA from Urine
3. Materials and Methods
3.1. Reagents and Materials
3.2. Chromatographic Conditions
3.3. Synthesis
3.4. Polymer Synthesis: MIP and NIP
3.5. Characterization of CDHB crystals
3.6. Characterization of Polymers
3.6.1. Physicochemical Characterization of Polymers
3.6.2. Adsorption Studies
3.6.3. Binding Analysis of Molecularly-Imprinted Polymers
3.7. Urine Sample Preparation
3.8. Validation Procedure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hewitt, T.C.; Flack, C.L.; Kolodziejczyk, J.K.; Chacon, A.M.; D’Ovidio, K.L. Occurrence of zearalenone in fresh corn and corn products collected from local Hispanic markets in San Diego County, CA. Food Control 2012, 26, 300–304. [Google Scholar] [CrossRef]
- Urraca, J.L.; Marazuela, M.D.; Merino, E.R.; Orellana, G.; Moreno-Bondi, M.C. Molecularly imprinted polymers with a streamlined mimic for zearalenone analysis. J Chromatogr. A 2006, 1116, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Kuiper-Goodman, T.; Scott, P.M.; Watanabe, H. Risk assessment of the mycotoxin zearalenone. Rev. Toxicol. Pharmacol. 1987, 7, 253–306. [Google Scholar] [CrossRef]
- Haggler, W.M., Jr.; Towers, N.R.; Mirocha, C.J.; Eppley, R.M.; Bryden, W.L. Zearalenone: Mycotoxin or mycoestrogen? In Fusarium: Paul E. Nelson Memorial Symposium; Summerell, B.A., Leslie, J.F., Bachouse, D., Bryden, W.L., Burges, L.W., Eds.; APS Press: St. Paul, MN, USA, 2001; pp. 321–331. [Google Scholar]
- Minervini, F.; Dell’Aquila, M.E. Zearalenone and reproductive function in farm animals. Int. J. Mol. Sci. 2008, 9, 2570–2584. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, J.; Miturski, R.; Semczuk, A.; Kotarski, J.; Jakowicki, J. Tissue zearalenone concentration in normal, hyperplastic and neoplastic human endometrium. Ginekol. Pol. 1998, 69, 363–366. (In Polish) [Google Scholar]
- Faber, K.A.; Hughes, C., Jr. The effect of neonatal exposure to diethylstilbestrol, genistein, and zearalenone on pituitary responsiveness and sexually dimorphic nucleus volume in the castrated adult rat. Biol. Reprod. 1991, 45, 649–653. [Google Scholar] [CrossRef]
- Hilakivi-Clarke, L.; Onojafe, I.; Raygada, M.; Cho, E.; Skaar, T.; Russo, I.; Clarke, R. Prepubertal exposure to zearalenone or genistein reduces mammary tumorigenesis. Br. J. Cancer 1999, 80, 1682–1688. [Google Scholar] [CrossRef]
- Thouvenot, D.; Morfin, R.F. Interferences of zearalenone, zearalanol or estradiol-17β with the steroid-metabolizing enzymes of the human prostate gland. J. Steroid Biochem. 1980, 13, 1337–1345. [Google Scholar] [CrossRef]
- Häggblom, P.; Nordkvist, E. Deoxynivalenol, zearalenone, and Fusarium graminearum contamination of cereal straw; field distribution; and sampling of big bales. Mycotoxin. Res. 2015, 31, 101–107. [Google Scholar] [CrossRef]
- Thanner, S.; Czeglédi, L.; Schwartz-Zimmermann, H.E.; Berthiller, F.; Gutzwiller, A. Urinary deoxynivalenol (DON) and zearalenone (ZEA) as biomarkers of DON and ZEA exposure of pigs. Mycotoxin Res. 2016, 32, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Cirlini, M.; Mazzeo, T.; Roncoroni, L.; Lombardo, V.; Elli, L.; Bardella, M.T.; Agostoni, C.; Doneda, L.; Brighenti, F.; Dall’Asta, C.; et al. Are Treated Celiac Patients at Risk for Mycotoxins? An Italian Case-Study. Toxins 2017, 9, 11. [Google Scholar] [CrossRef]
- Nováková, L. Advances in sample preparation for biological fluids. LC-GC 2016, 29, 9–15. [Google Scholar]
- Szultka, M.; Buszewski, B. Past, present, and future of solid phase extraction: A review. Crit. Rev. Anal. Chem. 2012, 42, 198–213. [Google Scholar]
- Escrivá, L.; Manyes, L.; Font, G.; Berrada, H. Mycotoxin Analysis of Human Urine by LC-MS/MS: A Comparative Extraction Study. Toxins 2017, 9, 330. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Carrasco, Y.; Moltó, J.C.; Mañes, J.; Berrada, H. Development of microextraction techniques in combination with GC–MS/MS for the determination of mycotoxins and metabolitesin human urine. J. Sep. Sci. 2017, 40, 1572–1582. [Google Scholar] [CrossRef]
- Belhassen, H.; Jiménez-Díaz, I.; Arrebola, J.P.; Ghali, R.; Ghorbel, H.; Olea, N.; Hedili, A. Zearalenone and its metabolites in urine and breast cancer risk: A case-control study in Tunisia. Chemosphere 2015, 128, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Songsermsakul, P.; Böhm, J.; Aurich, C.; Zentek, J.; Razzazi-Fazeli, E. The levels of zearalenone and its metabolites in plasma, urine and faeces of horses fed with naturally, Fusarium toxin-contaminated oats. J. Anim. Physiol. Anim. Nutr. 2013, 97, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Jodlbauer, J.; Zöllner, P.; Lindner, W. Determination of zearalenone and its metabolites in urine and tissue samples of cow and pig by LC-MS/MS. Mycotoxin Res. 2000, 16, 174–178. [Google Scholar] [CrossRef]
- Songsermsakul, P.; Sontag, G.; Cichna-Markl, M.; Zentek, J.; Razzazi-Fazeli, E. Determination of zearalenone and its metabolites in urine, plasma and faeces of horses by HPLC–APCI–MS. J. Chromatogr. B 2006, 843, 252–261. [Google Scholar] [CrossRef]
- Matraszek-Zuchowska, I.; Wozniak, B.; Zmudzki, J. Determination of zeranol, taleranol, zearalanone, α-zearalenol, β-zearalenol and zearalenone in urine by LC-MS/MS. Food Addit. Contam. Part A 2013, 30, 987–994. [Google Scholar] [CrossRef]
- Heyndrickx, E.; Sioen, I.; Huybrechts, B.; Callebaut, A.; De Henauw, S.; De Saeger, S. Human biomonitoring of multiple mycotoxins in the Belgian population: Results of the BIOMYCO study. Environ. Int. 2015, 84, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Fleck, S.C.; Churchwell, M.I.; Doerge, D.R.; Teeguarden, J.G. Urine and serum biomonitoring of exposure to environmental estrogens II: Soy isoflavones and zearalenone in pregnant women. Food Chem. Toxicol. 2016, 95, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Takagi, M.; Uno, S.; Kokushi, E.; Sato, F.; Wijayagunawardane, M.; Fink-Gremmels, J. Measurement of urinary concentrations of the mycotoxins zearalenone and sterigmatocystin as biomarkers of exposure in mares. Reprod. Domest. Anim. 2018, 53, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Bojdi, M.K.; Mashhadizadeh, M.H.; Behbahani, M.; Farahani, A.; Davarani, S.S.H. Bagheri, A. Synthesis, characterization and application of novel lead imprinted polymer nanoparticles as a high selective electrochemical sensor for ultra-trace determination of lead ions in complex matrixes spiked environmental water samples. Electrochim. Acta 2014, 136, 59–65. [Google Scholar] [CrossRef]
- Bojdi, M.K.; Behbahani, M.; Sahragard, A.; Amin, B.G.; Fakhari, A.; Bagheri, A. A palladium imprinted polymer for highly selective and sensitive electrochemical determination of ultra-trace of palladium ions. Electrochim. Acta 2014, 149, 108–116. [Google Scholar] [CrossRef]
- Urraca, J.L.; Carbajo, M.C.; Torralvo, M.J.; González-Vázquez, J.; Orellana, G.; Moreno-Bondi, M.C. Effect of the template and functional monomer on the textural properties of molecularly imprinted polymers. Biosens. Bioelectron. 2008, 24, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Villoslada, F.; Urraca, J.L.; Moreno-Bondi, M.C.; Orellan, G. Zearalenone sensing with molecularly imprinted polymers and tailored fluorescent probes. Sens. Actuators B Chem. 2007, 121, 67–73. [Google Scholar] [CrossRef]
- Gadzała-Kopciuch, R.; Cendrowski, K.; Cesarz, A.; Kiełbasa, P.; Buszewski, B. Determination of zearalenone and its metabolites in endometrial cancer by coupled separation techniques. Anal. Bioanal. Chem. 2011, 401, 2069–2078. [Google Scholar] [CrossRef]
- Kwaśniewska, K.; Gadzała-Kopciuch, R.; Cendrowski, K. Analytical procedure for the determination of zearalenone in environmental and biological samples. Crit. Rev. Anal. Chem. 2015, 45, 119–130. [Google Scholar] [CrossRef]
- Frisch, G.W.; Trucks, H.B.; Schlegel, G.E.; Scuseria, M.A.; Robb, J.R.; Cheeseman, G.; Scalmani, V.; Barone, B.; Mennucci, G.A.; Petersson, H.; et al. Gaussian09; Gaussian. Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Stewart, J.J. MOPAC Manual. A General Molecular Orbital Package, DTIC Document. Available online: http://OpenMOPAC.net/Manual (accessed on 1 October 2017).
- Koppen, R.; Riedel, J.; Emmerling, F.; Kocha, M. (3S, 11Z)-14, 16-Dihydroxy-3-methyl-3, 4, 5, 6, 9, 10-hexahydro-1H-2-benzoxacyclotetradecine-1, 7 (8H)-dione (cis-zearalenone): A redetermination. Acta Crystallogr. Sect. E Struct. Rep. Online 2012, 68, o832. [Google Scholar] [CrossRef]
- Zhao, L.-L.; Gai, Y.; Kobayashi, H.; Hua, C.-Q.; Zhanga, H.-P. (4S, 8S, 9R, 12E)-8, 9, 16, 18-Tetrahydroxy-4-methyl-3-oxabicyclo [12.4. 0] octadeca-12, 14, 16, 18-tetraen-2-one monohydrate. Acta Crystallogr. Sect. E Struct. Rep. Online 2008, 64, o999. [Google Scholar] [CrossRef] [PubMed]
- Allen, F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. B: Struct. Sci. 2002, 58, 380–388. [Google Scholar] [CrossRef]
- Sathesh, V.; Umamahesh, B.; Ramachandran, G.; Rathore, R.S.; Sathiyanarayanan, K.I. Direct anti and regio-specific aldol reactions of cyclododecanone catalyzed by alkali metal hydroxides: Implications for supramolecular helical design. New J. Chem. 2012, 36, 2292–2301. [Google Scholar] [CrossRef]
- Lemmerer, A. Two-dimensional layers using different combinations of hydrogen bonded rings in three ammonium carboxylate salts. J. Chem. Crystallogr. 2012, 42, 338–344. [Google Scholar] [CrossRef]
- Yusof, N.A.; Rahman, S.K.A.; Hussein, M.Z.; Ibrahim, N.A. Preparation and characterization of molecularly imprinted polymer as SPE sorbent for melamine isolation. Polymers 2013, 5, 1215–1228. [Google Scholar] [CrossRef]
- Preira da Silva, C.; Soares Emídio, E.; Rodrigues de Marchi, M.R. Method validation using weighted linear regression models for quantification of UV filters in water samples. Talanta 2015, 131, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Economou, A.; Botitsi, H.; Antoniou, S.; Tsipi, D. Determination of multi-class pesticides in wines by solid-phase extraction and liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2009, 1216, 5856–5867. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A Short History of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Row, K.H. Characteristic and synthetic approach of molecularly imprinted polymer. Int. J. Mol. Sci. 2006, 7, 155–178. [Google Scholar] [CrossRef]
- Vasapollo, G.; Del Sole, R.; Mergola, L.; Lazzoi, M.R.; Scardino, A.; Scorrano, S.; Mele, G. Molecularly imprinted polymers: Present and future prospective. Int. J. Mol. Sci. 2011, 12, 5908–5945. [Google Scholar] [CrossRef]
- Wang, H.L.; Fu, W.; Shen, Y.; Tan, H.; Xu, H. Molecularly Imprinted polymers for selective extraction of Oblongifolin C from Garcinia yunnanensis. Molecules 2017, 22, 508. [Google Scholar] [CrossRef] [PubMed]
- International Conference on Harmonization (ICH). Q2A: Text on Validation of Analytical Procedures, March 1995. Available online: https://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Quality/Q2_R1/Step4/Q2_R1_Guideline.pdf (accessed on 27 March 2019).
- International Conference on Harmonization (ICH). Q2B: Validation of Analytical Procedures: Methodology, May 1997. 19 May. Available online: https://www.fda.gov/downloads/drugs/guidances/ucm073384.pdf (accessed on 27 March 2019).
- U.S. FDA. Guidance for Industry: Analytical Procedures and Methods Validation: Chemistry, Manufacturing and Controls Documentation. July 2015. Available online: https://www.fda.gov/downloads/drugs/guidances/ucm386366.pdf (accessed on 27 March 2019).













| Adsorbent | Recovery (SD) (%) | Methods | LOD/LOQ | Concentration Range | Subjects | Ref. |
|---|---|---|---|---|---|---|
| ELISA | not specified | LC-MS/MS | 1)0.02/0.007 mg·kg−1 | 0.3–100 ng·mL−1 | Pigs | [11] |
| RP C18 Phenomenex | 96.6 (3.8) | LC-MS/MS | 0.1/0.5 pg | 0.5–100 pg | Cow, pigs | [19] |
| ISOLUTE® C18/immunoaffinity column Easi-Extract® Zearalenone | 108.2 (7.5) | HPLC–APCI–MS | 0.1/0.5 μg·L−1 | 0.5–100 μg·L−1 | Horse | [20] |
| 2)LLE/BakerBond C18 and NH2 | 94.3–114.0 (9–19.8) | LC-MS/MS | Detection limit: 0.11 μg·L−1 | 0.18–5 μg·L−1 | Animal | [21] |
| SAX SPE cartridge | not specified | LC-MS/MS | LOD: 61 pg·mL−1 α-ZEL; 117 pg·mL−1 β-ZEL14GlcA | - | Human | [22] |
| Supelco Titan C18 | not specified | UHPLC-MS/MS | LOD: 0.31 μg·L–1 ZEA; 0.11 μg·L−1 α-ZEL | - | Pregnant women | [23] |
| C18 | 94.3 | LC-ESI/MS/MS | 0.02/0.05 ng·ml−1 | 1.81 pg·mg−1 Creatinine | Mares | [24] |
| MIP-CDHB | 95.2–98.2 | HPLC-FLD | 3)LOQ: 5.4 ng·mL−1 | 10–1000 ng·mL−1 | Lyophilized human urine | [this work] |
| (1.5–2.0) | ||||||
| MIP-ZEA | 94.1–97.1 | 6.3 ng·mL−1 | ||||
| (1.6–1.9) 4) | ||||||
| NIP | 72.1–80.0 | 36.9 ng·mL−1 | 50–1000 ng·mL−1 | |||
| (8.2–11.2) 4) | ||||||
| ImmunoClean C ZON | 86.5–92.5 | 10.8 ng·mL−1 | 15–500 ng·mL−1 | |||
| (2.3–5.2) 4) | ||||||
| ImmunoClean C+ ZON | 88.2–92.1 | 8.3 ng·mL−1 | ||||
| (1.9–3.8) 4) |
| Wavenumber (cm−1) | Frequency Assignment |
|---|---|
| 3402 | O-H stretching, and possibly intra molecular hydrogen bonded –OH groups |
| 3200 | O-H stretching, and possibly intra molecular hydrogen bonded –OH groups |
| 2953 | C-H stretching |
| 1686 | C=O stretching |
| 1469 | C-C stretching in aromatic ring |
| 1255 | C-O stretching |
| 850 | C-H out of plane |
| Conformer | Energy (kcal/mol, [au]) | Dipole Moment (Debye) | φ1 (°) | EXP φ1 (°) | φ3 (°) | EXP φ3 (°) |
|---|---|---|---|---|---|---|
| K1 | 0.00 [−1041.672265] | 2.00 | 179.97 | −172.1 | −149.53 | −138.96 |
| K2 | 3.28 | 1.10 | 0.18 | 6.03 | −149.44 | −139.96 |
| K3 | 4.98 | 4.05 | 179.92 | −172.1 | −83.79 | −78.63 |
| K4 | 7.85 | 1.50 | 0.56 | 6.03 | −83.79 | −78.63 |
| D-H | A | d(D-H) | d(H...A) | d(D...A) | <(DHA) | |
|---|---|---|---|---|---|---|
| Mol 1 | O1-H101 | O3 | 0.82 | 1.9 | 2.6218(17) | 145.3 |
| O1W-H1W1 | O3[x−1/2, −y+3/2, z−1/2] | 0.95 | 1.90 | 2.8449(18) | 174.2 | |
| O2-H1O2 | O23[−x, −y+1, −z] | 0.82 | 1.96 | 2.7820(18) | 175.5 | |
| Mol 2 | O21-H21A_a | O23 | 0.82 | 1.93 | 2.635(2) | 143.4 |
| O21B-H21B_b | O24 | 0.82 | 1.83 | 2.528(5) | 141.9 | |
| O22-H22O | O1W | 0.82 | 1.84 | 2.6596(19) | 174.8 | |
| O1W-H2W1 | O22[−x−3/2, y+1/2, −z−1/2] | 0.94 | 1.91 | 2.8393(17) | 168.7 |
| Type of SPE Sorbent | Range of Concentration (ng mL−1) | Calibration Equation y = mx ± b | r2 | LOD (ng mL−1) | LOQ (ng mL−1) | % ME |
|---|---|---|---|---|---|---|
| MIP-CDHB | 10–1000 | y = 0.0254x + 0.6721 | 0.9990 | 1.8 | 5.4 | 15.1% |
| MIP-ZEA | y = 0.0240x + 0.4383 | 0.9985 | 2.1 | 6.3 | 19.7% | |
| NIP | 50–1000 | y = 0.0185x + 1.7157 | 0.9815 | 11.2 | 36.9 | 38.1% |
| ImmunoClean C ZON | 15–500 *) | y = 0.0226x – 0.0549 | 0.9918 | 3.2 | 10.8 | 24.4% |
| ImmunoClean C+ ZON | y = 0.0241x + 0.8189 | 0.9990 | 2.5 | 8.3 | 19.4% |
| Type of SPE Sorbent | t/qe = const∙t | r2 | qe |
|---|---|---|---|
| MIP-CDHB | t/qe = 2.2454·t | 0.999 | 1.8 |
| MIP-ZEA | t/qe = 2.1311·t | 0.999 | 2.1 |
| Sorbent | ZEA Concentration | |||||||
|---|---|---|---|---|---|---|---|---|
| 20 ng·mL−1 | 100 ng·mL−1 | 400 ng·mL−1 | 500 ng·mL−1 | |||||
| R (%) | RSD (%) | R (%) | RSD (%) | R (%) | RSD (%) | R (%) | RSD (%) | |
| MIP-CDHB | 98.2 | 1.5 | 97.3 | 1.9 | 95.2 | 2.0 | 96.5 | 1,8 |
| MIP-ZEA | 97.1 | 1.6 | 96.5 | 1.7 | 94.1 | 1.9 | 95.1 | 1.9 |
| NIP | 72.1 | 9.6 | 80.0 | 8.2 | 78.1 | 10.3 | 75.2 | 11.2 |
| ImmunoClean C ZON | 90.0 | 4.5 | 92.5 | 2.3 | 90.1 | 4.3 | 86.5 | 5.2 |
| ImmunoClean C+ ZON | 91.8 | 3.2 | 92.1 | 1.9 | 91.4 | 2.2 | 88.2 | 3.8 |
| Empirical Formula | C38H56O9 |
|---|---|
| Formula weight | 656.83 |
| Temperature; K | 293(2) |
| Wavelength; Å | 0.71073 |
| Crystal system, space group | Monoclinic, P21/n |
| Unit cell dimensions; Å and ° | a = 17.1943(7) |
| b = 8.1870(4) | |
| c = 25.8661(11) | |
| beta = 92.282(4) | |
| Volume; Å3 | 3638.3(3) |
| Z, Calculated density; Mg/m3 | 4, 1.199 |
| Absorption coefficient; mm−1 | 0.084 |
| F(000) | 1424 |
| Crystal size; mm | 0.45 × 0.26 × 0.13 |
| Theta range for data collection | 2.37 to 28.15º |
| Limiting indices | −22 ≤ h ≤ 22, −9 ≤ k ≤ 10, −34 ≤ l ≤ 31 |
| Reflections collected/unique | 23663/7895 [R(int) = 0.0524] |
| Completeness to theta | 26.00 99.9% |
| Max. and min. transmission | 0.9902 and 0.9667 |
| Refinement method | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 7895/0/434 |
| Goodness-of-fit on F2 | 0.779 |
| Final R indices [I > 2sigma(I)] | R1 = 0.0420, wR2 = 0.0805 |
| R indices (all data) | R1 = 0.1415, wR2 = 0.0978 |
| Largest diff. peak and hole; e.A−3 | 0.214 and −0.147 |
| Polymer Code | SBET (m2·g−1) | Vp (cm3·g−1) | dp (nm) |
|---|---|---|---|
| NIP | 182.24 ± 3.24 | 0.396 | 7.62 |
| MIP-CDHB | 247.52 ± 2.12 | 0.495 | 10.15 |
| MIP-ZEA | 251.25 ± 1.96 | 0.504 | 11.24 |
| Polymer Code | QMIP | QNIP | α |
|---|---|---|---|
| NIP | - | 38.18 | |
| MIP-CBHB | 48.40 | - | 1.27 |
| MIP-ZEA | 47.85 | - | 1.25 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gadzała-Kopciuch, R.; Kwaśniewska, K.; Ludwiczak, A.; Skrzyniarz, P.; Jakubowski, R.; Nowak, W.; Wojtczak, A.; Buszewski, B. Towards A New Approach for the Description of Cyclo–2,4-Dihydroxybenzoate, A Substance Which Effectively Mimics Zearalenone in Imprinted Polymers Designed for Analyzing Selected Mycotoxins in Urine. Int. J. Mol. Sci. 2019, 20, 1588. https://doi.org/10.3390/ijms20071588
Gadzała-Kopciuch R, Kwaśniewska K, Ludwiczak A, Skrzyniarz P, Jakubowski R, Nowak W, Wojtczak A, Buszewski B. Towards A New Approach for the Description of Cyclo–2,4-Dihydroxybenzoate, A Substance Which Effectively Mimics Zearalenone in Imprinted Polymers Designed for Analyzing Selected Mycotoxins in Urine. International Journal of Molecular Sciences. 2019; 20(7):1588. https://doi.org/10.3390/ijms20071588
Chicago/Turabian StyleGadzała-Kopciuch, Renata, Katarzyna Kwaśniewska, Agnieszka Ludwiczak, Piotr Skrzyniarz, Rafał Jakubowski, Wiesław Nowak, Andrzej Wojtczak, and Bogusław Buszewski. 2019. "Towards A New Approach for the Description of Cyclo–2,4-Dihydroxybenzoate, A Substance Which Effectively Mimics Zearalenone in Imprinted Polymers Designed for Analyzing Selected Mycotoxins in Urine" International Journal of Molecular Sciences 20, no. 7: 1588. https://doi.org/10.3390/ijms20071588