TGF-β Signaling and the Epithelial-Mesenchymal Transition during Palatal Fusion
Abstract
:1. Introduction
2. The Role of the TGF-β Signaling Pathway in Palatal Fusion
2.1. MEE Cell Fate Includes Program Cells Death, Cell Migration and Epithelial-Mesenchyme Transition
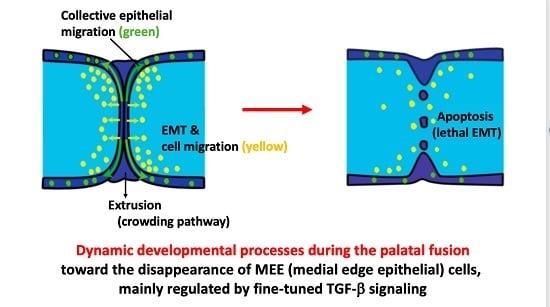
2.2. Epithelial Migration, Extrusion, and Apoptosis at the MES
2.2.1. Epithelial (MEE) Cell Migration
2.2.2. Extrusion
2.2.3. Apoptosis
2.3. Human Syndromes with Palatal Defects Related to TGF-β Signaling
2.4. Expression of TGF-βs in the Palate and the Resulting Phenotypes When Genes Related to TGF-βs Are Deleted
2.5. Palatal Development and Expression of TGF-β Receptors (TβRs)
2.6. Smad-Dependent Signaling Pathway
2.7. Non-Smad Signaling Pathways
3. Other Signaling Pathways and Possible Cross-Talks with TGF-β Signaling during Palatal Development
3.1. BMP Signaling
3.2. FGF Signaling
3.3. Ephrin
3.4. Wnt Signaling
3.5. Extracellular Matrix (ECM)
4. Conclusion and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Mossey, P.A.; Little, J.; Munger, R.G.; Dixon, M.J.; Shaw, W.C. Cleft lip and palate. Lancet 2009, 374, 1773–1785. [Google Scholar] [CrossRef]
- Ferguson, M.W. Palate development. Development 1988, 103, 41–60. [Google Scholar] [PubMed]
- Vaziri Sani, F.; Hallberg, K.; Harfe, B.D.; McMahon, A.P.; Linde, A.; Gritli-Linde, A. Fate-mapping of the epithelial seam during palatal fusion rules out epithelial-mesenchymal transformation. Dev. Biol. 2005, 285, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Mori, C.; Nakamura, N.; Okamoto, Y.; Osawa, M.; Shiota, K. Cytochemical identification of programmed cell death in the fusing fetal mouse palate by specific labelling of DNA fragmentation. Anat. Embryol. (Berl.) 1994, 190, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, R.; Valencia, C.; Chandraratna, R.A.; Covarrubias, L. Programmed cell death is required for palate shelf fusion and is regulated by retinoic acid. Dev. Biol. 2002, 245, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, R.; Covarrubias, L. Death is the major fate of medial edge epithelial cells and the cause of basal lamina degradation during palatogenesis. Development 2004, 131, 15–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, J.; Suzuki, A.; Yokota, T.; Ho, T.V.; Pelikan, R.; Urata, M.; Sanchez-Lara, P.A.; Chai, Y. TGF-β regulates epithelial-mesenchymal interactions through WNT signaling activity to control muscle development in the soft palate. Development 2014, 141, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.Z.; Ding, J. Analysis of cell migration, transdifferentiation and apoptosis during mouse secondary palate fusion. Development 2006, 133, 3341–3347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Yokota, T.; Iwata, J.; Chai, Y. TGF-β -mediated FasL-Fas-Caspase pathway is crucial during palatogenesis. J. Dent. Res. 2011, 90, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Alvarez, C.; Tudela, C.; Perez-Miguelsanz, J.; O’Kane, S.; Puerta, J.; Ferguson, M.W. Medial edge epithelial cell fate during palatal fusion. Dev. Biol. 2000, 220, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Carette, M.J.; Ferguson, M.W. The fate of medial edge epithelial cells during palatal fusion in vitro: An analysis by DiI labelling and confocal microscopy. Development 1992, 114, 379–388. [Google Scholar] [PubMed]
- Ferguson, W.J. Epithelial-mesenchymal interactions during vertebrate palatogenesis. Curr. Top. Dev. Biol. 1984, 19, 137–164. [Google Scholar] [PubMed]
- Fitchett, J.E.; Hay, E.D. Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fuse. Dev. Biol. 1989, 131, 455–474. [Google Scholar] [CrossRef]
- Shuler, C.F.; Guo, Y.; Majumder, A.; Luo, R.Y. Molecular and morphologic changes during the epithelial-mesenchymal transformation of palatal shelf medial edge epithelium in vitro. Int. J. Dev. Biol. 1991, 35, 463–472. [Google Scholar] [PubMed]
- Shuler, C.F.; Halpern, D.E.; Guo, Y.; Sank, A.C. Medial edge epithelium fate traced by cell lineage analysis during epithelial-mesenchymal transformation in vivo. Dev. Biol. 1992, 154, 318–330. [Google Scholar] [CrossRef]
- Griffith, C.M.; Hay, E.D. Epithelial-mesenchymal transformation during palatal fusion: Carboxyfluorescein traces cells at light and electron microscopic levels. Development 1992, 116, 1087–1099. [Google Scholar] [PubMed]
- Kang, P.; Svoboda, K.K. Epithelial-mesenchymal transformation during craniofacial development. J. Dent. Res. 2005, 84, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Gritli-Linde, A. Molecular control of secondary palate development. Dev. Biol. 2007, 301, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Nawshad, A. Complexity in interpretation of embryonic epithelial-mesenchymal transition in response to transforming growth factor-β signaling. Cells Tissues Organs 2007, 185, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Liu, C.C.; Nawshad, A. Mechanisms of palatal epithelial seam disintegration by transforming growth factor (TGF) β3. Dev. Biol. 2007, 309, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Tanaka, E.; Ito, Y.; Maeno, M.; Iwata, K.; Shimizu, N.; Shuler, C.F. The expression of TGF-β3 for epithelial-mesenchyme transdifferentiated MEE in palatogenesis. J. Mol. Histol. 2010, 41, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Jalali, A.; Zhu, X.; Liu, C.; Nawshad, A. Induction of palate epithelial mesenchymal transition by transforming growth factor β3 signaling. Dev. Growth Differ. 2012, 54, 633–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proetzel, G.; Pawlowski, S.A.; Wiles, M.V.; Yin, M.; Boivin, G.P.; Howles, P.N.; Ding, J.; Ferguson, M.W.; Doetschman, T. Transforming growth factor-β 3 is required for secondary palate fusion. Nat. Genet. 1995, 11, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Kaartinen, V.; Voncken, J.W.; Shuler, C.; Warburton, D.; Bu, D.; Heisterkamp, N.; Groffen, J. Abnormal lung development and cleft palate in mice lacking TGF-β3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 1995, 11, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. TGF-β signal transduction. Annu. Rev. Biochem. 1998, 67, 753–791. [Google Scholar] [CrossRef] [PubMed]
- Kaartinen, V.; Cui, X.M.; Heisterkamp, N.; Groffen, J.; Shuler, C.F. Transforming growth factor-β3 regulates transdifferentiation of medial edge epithelium during palatal fusion and associated degradation of the basement membrane. Dev. Dyn. 1997, 209, 255–260. [Google Scholar] [CrossRef] [Green Version]
- Taya, Y.; O’Kane, S.; Ferguson, M.W. Pathogenesis of cleft palate in TGF-β3 knockout mice. Development 1999, 126, 3869–3879. [Google Scholar] [PubMed]
- Martinez-Sanz, E.; Del Rio, A.; Barrio, C.; Murillo, J.; Maldonado, E.; Garcillan, B.; Amoros, M.; Fuerte, T.; Fernandez, A.; Trinidad, E.; et al. Alteration of medial-edge epithelium cell adhesion in two Tgf-β3 null mouse strains. Differentiation 2008, 76, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Larsson, J.; Goumans, M.J.; Sjostrand, L.J.; van Rooijen, M.A.; Ward, D.; Leveen, P.; Xu, X.; ten Dijke, P.; Mummery, C.L.; Karlsson, S. Abnormal angiogenesis but intact hematopoietic potential in TGF-β type I receptor-deficient mice. EMBO J. 2001, 20, 1663–1673. [Google Scholar] [CrossRef] [PubMed]
- Oshima, M.; Oshima, H.; Taketo, M.M. TGF-β receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev. Biol. 1996, 179, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Yeo, J.Y.; Chytil, A.; Han, J.; Bringas, P., Jr.; Nakajima, A.; Shuler, C.F.; Moses, H.L.; Chai, Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 2003, 130, 5269–5280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, X.M.; Shuler, C.F. The TGF-β type III receptor is localized to the medial edge epithelium during palatal fusion. Int. J. Dev. Biol. 2000, 44, 397–402. [Google Scholar] [PubMed]
- Nakajima, A.; Ito, Y.; Asano, M.; Maeno, M.; Iwata, K.; Mitsui, N.; Shimizu, N.; Cui, X.M.; Shuler, C.F. Functional role of transforming growth factor-β type III receptor during palatal fusion. Dev. Dyn. 2007, 236, 791–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, J.; Parada, C.; Chai, Y. The mechanism of TGF-β signaling during palate development. Oral Dis. 2011, 17, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Ito, Y.; Tanaka, E.; Sano, R.; Karasawa, Y.; Maeno, M.; Iwata, K.; Shimizu, N.; Shuler, C.F. Functional role of TGF-β receptors during palatal fusion in vitro. Arch. Oral Biol. 2014, 59, 1192–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Han, J.; Ito, Y.; Bringas, P., Jr.; Deng, C.; Chai, Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-β/BMP signaling during tooth and palate development. Dev. Cell 2008, 15, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Parada, C.; Chai, Y. Roles of BMP signaling pathway in lip and palate development. Front. Oral Biol. 2012, 16, 60–70. [Google Scholar] [PubMed]
- Yuan, G.; Zhan, Y.; Gou, X.; Chen, Y.; Yang, G. TGF-β signaling inhibits canonical BMP signaling pathway during palate development. Cell Tissue Res. 2018, 371, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Stanier, P.; Pauws, E. Development of the lip and palate: FGF signaling. Front. Oral Biol. 2012, 16, 71–80. [Google Scholar] [PubMed]
- Rice, R.; Spencer-Dene, B.; Connor, E.C.; Gritli-Linde, A.; McMahon, A.P.; Dickson, C.; Thesleff, I.; Rice, D.P. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Investig. 2004, 113, 1692–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, M.; Chen, Z.; Xiao, Q.; Li, R.; Chen, Z. A review of FGF signaling in palate development. Biomed. Pharmacother. 2018, 103, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.D.; Serrano, M.J. Ephrin regulation of palate development. Front. Physiol. 2012, 3, 376. [Google Scholar] [CrossRef] [PubMed]
- Blavier, L.; Lazaryev, A.; Groffen, J.; Heisterkamp, N.; DeClerck, Y.A.; Kaartinen, V. TGF-β3-induced palatogenesis requires matrix metalloproteinases. Mol. Biol. Cell 2001, 12, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Kitase, Y.; Yamashiro, K.; Fu, K.; Richman, J.M.; Shuler, C.F. Spatiotemporal localization of periostin and its potential role in epithelial-mesenchymal transition during palatal fusion. Cells Tissues Organs 2011, 193, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Levi, B.; Brugman, S.; Wong, V.W.; Grova, M.; Longaker, M.T.; Wan, D.C. Palatogenesis: Engineering, pathways and pathologies. Organogenesis 2011, 7, 242–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Lewis, A.E.; Singh, V.; Ma, X.; Adelstein, R.; Bush, J.O. Convergence and extrusion are required for normal fusion of the mammalian secondary palate. PLoS Biol. 2015, 13, e1002122. [Google Scholar] [CrossRef] [PubMed]
- Massague, J.; Wotton, D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000, 19, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signaling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, A.; Thakur, N.; Grimsby, S.; Marcusson, A.; von Bulow, V.; Schuster, N.; Zhang, S.; Heldin, C.H.; Landstrom, M. The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 2008, 10, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Danielian, P.S.; Muccino, D.; Rowitch, D.H.; Michael, S.K.; McMahon, A.P. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998, 8, 1323–1326. [Google Scholar] [CrossRef]
- Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999, 21, 70–71. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Jiang, X.; Ito, Y.; Bringas, P., Jr.; Han, J.; Rowitch, D.H.; Soriano, P.; McMahon, A.P.; Sucov, H.M. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 2000, 127, 1671–1679. [Google Scholar] [PubMed]
- Kim, S.; Prochazka, J.; Bush, J.O. Live Imaging of Mouse Secondary Palate Fusion. J. Vis. Exp. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, T.J.; Martin, P. Wound repair at a glance. J. Cell Sci. 2009, 122, 3209–3213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weijer, C.J. Collective cell migration in development. J. Cell Sci. 2009, 122, 3215–3223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilina, O.; Friedl, P. Mechanisms of collective cell migration at a glance. J. Cell Sci. 2009, 122, 3203–3208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Rosenblatt, J. New emerging roles for epithelial cell extrusion. Curr. Opin. Cell Biol. 2012, 24, 865–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenhoffer, G.T.; Loftus, P.D.; Yoshigi, M.; Otsuna, H.; Chien, C.B.; Morcos, P.A.; Rosenblatt, J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 2012, 484, 546–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, J.; Yumoto, K.; Azhar, M.; Ninomiya-Tsuji, J.; Inagaki, M.; Hu, Y.; Deng, C.X.; Kim, J.; Mishina, Y.; Kaartinen, V. Tak1, Smad4 and Trim33 redundantly mediate TGF-β3 signaling during palate development. Dev. Biol. 2015, 398, 231–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, J.; Suzuki, A.; Pelikan, R.C.; Ho, T.V.; Sanchez-Lara, P.A.; Urata, M.; Dixon, M.J.; Chai, Y. Smad4-Irf6 genetic interaction and TGFβ-mediated IRF6 signaling cascade are crucial for palatal fusion in mice. Development 2013, 140, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Huang, Y.H.; Chen, M.; Su, J.; Zou, Y.; Bardeesy, N.; Iacobuzio-Donahue, C.A.; Massague, J. TGF-β Tumor Suppression through a Lethal EMT. Cell 2016, 164, 1015–1030. [Google Scholar] [CrossRef] [PubMed]
- Seelan, R.S.; Mukhopadhyay, P.; Warner, D.R.; Webb, C.L.; Pisano, M.; Greene, R.M. Epigenetic regulation of Sox4 during palate development. Epigenomics 2013, 5, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Maitra, A. EMT: Matter of Life or Death? Cell 2016, 164, 840–842. [Google Scholar] [CrossRef] [PubMed]
- Bodo, M.; Baroni, T.; Carinci, F.; Becchetti, E.; Bellucci, C.; Pezzetti, F.; Conte, C.; Evangelisti, R.; Carinci, P. TGFβ isoforms and decorin gene expression are modified in fibroblasts obtained from non-syndromic cleft lip and palate subjects. J. Dent. Res. 1999, 78, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Carinci, F.; Scapoli, L.; Palmieri, A.; Zollino, I.; Pezzetti, F. Human genetic factors in nonsyndromic cleft lip and palate: An update. Int. J. Pediatr. Otorhinolaryngol. 2007, 71, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Stuppia, L.; Capogreco, M.; Marzo, G.; La Rovere, D.; Antonucci, I.; Gatta, V.; Palka, G.; Mortellaro, C.; Tete, S. Genetics of syndromic and nonsyndromic cleft lip and palate. J. Craniofac. Surg. 2011, 22, 1722–1726. [Google Scholar] [CrossRef] [PubMed]
- Poswillo, D. Otomandibular deformity: Pathogenesis as a guide to reconstruction. J. Maxillofac. Surg. 1974, 2, 64–72. [Google Scholar] [CrossRef]
- Robbins, A.; Zarate, Y.A.; Hartzell, L.D. Combined Tongue-Palate Fusion with Alveolar Bands in a Patient with Pierre Robin Sequence and Van der Woude Syndrome. Cleft Palate Craniofac. J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kousa, Y.A.; Fuller, E.; Schutte, B.C. IRF6 and AP2A Interaction Regulates Epidermal Development. J. Investig. Dermatol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Nowaczyk, M.J.; Irons, M.B. Smith-Lemli-Opitz syndrome: Phenotype, natural history, and epidemiology. Am. J. Med. Genet. C Semin. Med. Genet. 2012, 160C, 250–262. [Google Scholar] [CrossRef] [PubMed]
- McKusick, V.A. The defect in Marfan syndrome. Nature 1991, 352, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Neptune, E.R.; Frischmeyer, P.A.; Arking, D.E.; Myers, L.; Bunton, T.E.; Gayraud, B.; Ramirez, F.; Sakai, L.Y.; Dietz, H.C. Dysregulation of TGF-β activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 2003, 33, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Judge, D.P.; Dietz, H.C. Marfan’s syndrome. Lancet 2005, 366, 1965–1976. [Google Scholar] [CrossRef]
- Dietz, H.C.; Loeys, B.; Carta, L.; Ramirez, F. Recent progress towards a molecular understanding of Marfan syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2005, 139C, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Rommel, K.; Mishra, A.; Karck, M.; Haverich, A.; Schmidtke, J.; Arslan-Kirchner, M. TGFBR1 and TGFBR2 mutations in patients with features of Marfan syndrome and Loeys-Dietz syndrome. Hum. Mutat. 2006, 27, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Habashi, J.P.; Judge, D.P.; Holm, T.M.; Cohn, R.D.; Loeys, B.L.; Cooper, T.K.; Myers, L.; Klein, E.C.; Liu, G.; Calvi, C.; et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science 2006, 312, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Han, Y. Targeting TGF-β and the extracellular matrix in Marfan’s syndrome. Dev. Cell 2008, 15, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Loeys, B.L.; Chen, J.; Neptune, E.R.; Judge, D.P.; Podowski, M.; Holm, T.; Meyers, J.; Leitch, C.C.; Katsanis, N.; Sharifi, N.; et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet. 2005, 37, 275–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuguchi, T.; Collod-Beroud, G.; Akiyama, T.; Abifadel, M.; Harada, N.; Morisaki, T.; Allard, D.; Varret, M.; Claustres, M.; Morisaki, H.; et al. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat. Genet. 2004, 36, 855–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzpatrick, D.R.; Denhez, F.; Kondaiah, P.; Akhurst, R.J. Differential expression of TGF β isoforms in murine palatogenesis. Development 1990, 109, 585–595. [Google Scholar] [PubMed]
- Pelton, R.W.; Hogan, B.L.; Miller, D.A.; Moses, H.L. Differential expression of genes encoding TGFs β 1, β 2, and β 3 during murine palate formation. Dev. Biol. 1990, 141, 456–460. [Google Scholar] [CrossRef]
- Kulkarni, A.B.; Ward, J.M.; Yaswen, L.; Mackall, C.L.; Bauer, S.R.; Huh, C.G.; Gress, R.E.; Karlsson, S. Transforming growth factor-β 1 null mice. An animal model for inflammatory disorders. Am. J. Pathol. 1995, 146, 264–275. [Google Scholar] [PubMed]
- Mu, L.; Katsaros, D.; Lu, L.; Preti, M.; Durando, A.; Arisio, R.; Yu, H. TGF-β1 genotype and phenotype in breast cancer and their associations with IGFs and patient survival. Br. J. Cancer 2008, 99, 1357–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanford, L.P.; Ormsby, I.; Gittenberger-de Groot, A.C.; Sariola, H.; Friedman, R.; Boivin, G.P.; Cardell, E.L.; Doetschman, T. TGFβ2 knockout mice have multiple developmental defects that are non-overlapping with other TGFβ knockout phenotypes. Development 1997, 124, 2659–2670. [Google Scholar] [PubMed]
- Stenvers, K.L.; Tursky, M.L.; Harder, K.W.; Kountouri, N.; Amatayakul-Chantler, S.; Grail, D.; Small, C.; Weinberg, R.A.; Sizeland, A.M.; Zhu, H.J. Heart and liver defects and reduced transforming growth factor β2 sensitivity in transforming growth factor β type III receptor-deficient embryos. Mol. Cell. Biol. 2003, 23, 4371–4385. [Google Scholar] [CrossRef] [PubMed]
- Hilliard, S.A.; Yu, L.; Gu, S.; Zhang, Z.; Chen, Y.P. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J. Anat. 2005, 207, 655–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Mayo, J.; Xu, X.; Li, J.; Bringas, P., Jr.; Maas, R.L.; Rubenstein, J.L.; Chai, Y. Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development 2009, 136, 4225–4233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panopoulou, E.; Gillooly, D.J.; Wrana, J.L.; Zerial, M.; Stenmark, H.; Murphy, C.; Fotsis, T. Early endosomal regulation of Smad-dependent signaling in endothelial cells. J. Biol. Chem. 2002, 277, 18046–18052. [Google Scholar] [CrossRef] [PubMed]
- Dudas, M.; Nagy, A.; Laping, N.J.; Moustakas, A.; Kaartinen, V. Tgf-β3-induced palatal fusion is mediated by Alk-5/Smad pathway. Dev. Biol. 2004, 266, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.H.; Derynck, R. Specificity and versatility in tgf-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005, 21, 659–693. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.M.; Shiomi, N.; Chen, J.; Saito, T.; Yamamoto, T.; Ito, Y.; Bringas, P.; Chai, Y.; Shuler, C.F. Overexpression of Smad2 in Tgf-β3-null mutant mice rescues cleft palate. Dev. Biol. 2005, 278, 193–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pungchanchaikul, P.; Gelbier, M.; Ferretti, P.; Bloch-Zupan, A. Gene expression during palate fusion in vivo and in vitro. J. Dent. Res. 2005, 84, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Nawshad, A.; Medici, D.; Liu, C.C.; Hay, E.D. TGFβ3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J. Cell Sci. 2007, 120 Pt 9, 1646–1653. [Google Scholar] [CrossRef]
- Shiomi, N.; Cui, X.M.; Yamamoto, T.; Saito, T.; Shuler, C.F. Inhibition of SMAD2 expression prevents murine palatal fusion. Dev. Dyn. 2006, 235, 1785–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.E. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Fatyol, K.; Jin, C.; Wang, X.; Liu, Z.; Zhang, Y.E. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-β. Mol. Cell 2008, 31, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.B. The ever-increasing complexity of TGF-β signaling. Cytokine Growth Factor Rev. 2002, 13, 3–5. [Google Scholar] [CrossRef]
- Yu, L.; Hebert, M.C.; Zhang, Y.E. TGF-β receptor-activated p38 MAP kinase mediates Smad-independent TGF-β responses. EMBO J. 2002, 21, 3749–3759. [Google Scholar] [CrossRef] [PubMed]
- Lui, W.Y.; Lee, W.M.; Cheng, C.Y. Transforming growth factor β3 regulates the dynamics of Sertoli cell tight junctions via the p38 mitogen-activated protein kinase pathway. Biol. Reprod. 2003, 68, 1597–1612. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Ruest, L.B.; Svoboda, K.K. Regulation of epithelial-mesenchymal transition in palatal fusion. Exp. Biol. Med. (Maywood) 2009, 234, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Z.; Zhang, X.; Zhu, H.; Chen, Q.; Guo, M. The localization of adrenomedullin in rat kidney tissue and its inhibitory effect on the growth of cultured rat mesangial cells. Chin. Med. Sci. J. 2002, 17, 129–133. [Google Scholar] [PubMed]
- Graf, D.; Malik, Z.; Hayano, S.; Mishina, Y. Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev. 2016, 27, 129–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Song, Y.; Zhao, X.; Zhang, X.; Fermin, C.; Chen, Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development 2002, 129, 4135–4146. [Google Scholar] [PubMed]
- Charoenchaikorn, K.; Yokomizo, T.; Rice, D.P.; Honjo, T.; Matsuzaki, K.; Shintaku, Y.; Imai, Y.; Wakamatsu, A.; Takahashi, S.; Ito, Y.; et al. Runx1 is involved in the fusion of the primary and the secondary palatal shelves. Dev. Biol. 2009, 326, 392–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanai, J.; Chen, L.F.; Kanno, T.; Ohtani-Fujita, N.; Kim, W.Y.; Guo, W.H.; Imamura, T.; Ishidou, Y.; Fukuchi, M.; Shi, M.J.; et al. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J. Biol. Chem. 1999, 274, 31577–31582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Yasui, N.; Ito, K.; Huang, G.; Fujii, M.; Hanai, J.; Nogami, H.; Ochi, T.; Miyazono, K.; Ito, Y. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl. Acad. Sci. USA 2000, 97, 10549–10554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarper, S.E.; Kurosaka, H.; Inubushi, T.; Ono Minagi, H.; Kuremoto, K.I.; Sakai, T.; Taniuchi, I.; Yamashiro, T. Runx1-Stat3-Tgfb3 signaling network regulating the anterior palatal development. Sci. Rep. 2018, 8, 11208. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Popkie, A.P.; Xiong, W.; Li, L.; Wang, Y.; Phiel, C.J.; Chen, Y. Gsk3β is required in the epithelium for palatal elevation in mice. Dev. Dyn. 2010, 239, 3235–3246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, H.; Neubuser, A.; Kratochwil, K.; Balling, R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998, 12, 2735–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moustakas, A.; Heldin, C.H. Mechanisms of TGFβ-Induced Epithelial-Mesenchymal Transition. J. Clin. Med. 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Cui, X.M.; Shuler, C.F. Role of ERK1/2 signaling during EGF-induced inhibition of palatal fusion. Dev. Biol. 2003, 260, 512–521. [Google Scholar] [CrossRef]
- Del Rio, A.; Barrio, M.C.; Murillo, J.; Maldonado, E.; Lopez-Gordillo, Y.; Martinez-Sanz, E.; Martinez, M.L.; Martinez-Alvarez, C. Analysis of the presence of cell proliferation-related molecules in the Tgf-β3 null mutant mouse palate reveals misexpression of EGF and Msx-1. Cells Tissues Organs 2011, 193, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Girerd, B.; Montani, D.; Wang, X.J.; Galie, N.; Austin, E.D.; Elliott, G.; Asano, K.; Grunig, E.; Yan, Y.; et al. BMPR2 mutations and survival in pulmonary arterial hypertension: An individual participant data meta-analysis. Lancet Respir. Med. 2016, 4, 129–137. [Google Scholar] [CrossRef]







| Localization | Phenotype at Null Mutant | |
|---|---|---|
| TGF-β1 | MEE (prior to fuse) and Mesenchyme | (-) |
| TGF-β2 | MEE (prior to fuse) and Mesenchyme | 23% (+) |
| TGF-β3 | MEE | (+) |
| TβR1 | MEE | ? (Die at E10.5) |
| TβR2 | MEE | (+) |
| TβR3 | MEE | (-) |
| Fusion Stage | Vertical Growth | Elevation (Before Fuse) | Adhesion (Contact and Fusion) | After Fusion (MEE Disappear) | ||||
|---|---|---|---|---|---|---|---|---|
| Cells | Mesenchyme | Epithelium | Mesenchyme | Epithelium | Mesenchyme | Epithelium | Mesenchyme | Epithelium |
| Ligand | EphB2/B3 | pERK | TGFβ1/2 | TGFβ1/2 | BMP2/3/4 | TGFβ3 | BMP2/3/4 | |
| FGF7/10 | pMEK | EphB2/B3 | pERK | EphB2/B3 | BMP3 | Osr2 | ||
| Wnt5a | Shh | FGF10 | pMEK | FGF2/8/10/18 | FGF2/18 | |||
| FGFr1/2b | Shh | Shh | ||||||
| Wnt11 | Wnt11 | |||||||
| Smad2 | ||||||||
| pMEK | ||||||||
| Receptor | FGFr2 | FGFr1 | TβR1/2/3 | |||||
| FGFr2b | FGFr2 | |||||||
| Transcriptional | Snail | TBX1 | Msx1 | TBX1 | Msx1 | Snail | Snail | |
| Factor | TBX22 | TBX22 | Twist | Snail | TBX1 | |||
| Twist | Snail | Twist | Twist | |||||
| Msx1 | Runx1 | |||||||
| Pax9 | ||||||||
| Extracellular | MMP2/13 | MT-MMP | MMP2 | |||||
| Matrix | MMP13 | |||||||
| TIMP2 | ||||||||
| Periostin | ||||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakajima, A.; F. Shuler, C.; Gulka, A.O.D.; Hanai, J.-i. TGF-β Signaling and the Epithelial-Mesenchymal Transition during Palatal Fusion. Int. J. Mol. Sci. 2018, 19, 3638. https://doi.org/10.3390/ijms19113638
Nakajima A, F. Shuler C, Gulka AOD, Hanai J-i. TGF-β Signaling and the Epithelial-Mesenchymal Transition during Palatal Fusion. International Journal of Molecular Sciences. 2018; 19(11):3638. https://doi.org/10.3390/ijms19113638
Chicago/Turabian StyleNakajima, Akira, Charles F. Shuler, Alexander O. D. Gulka, and Jun-ichi Hanai. 2018. "TGF-β Signaling and the Epithelial-Mesenchymal Transition during Palatal Fusion" International Journal of Molecular Sciences 19, no. 11: 3638. https://doi.org/10.3390/ijms19113638