Mining the Plasma Cell Transcriptome for Novel Cell Surface Proteins
Abstract
:1. Introduction
2. Results
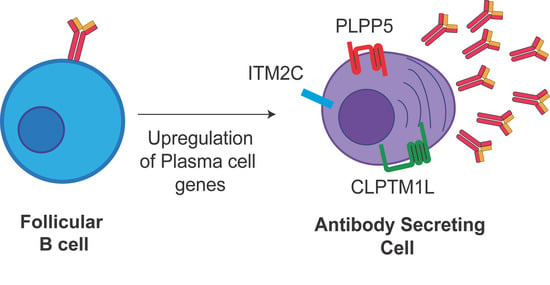
2.1. Identification of Candidate Cell Surface Proteins in Anibody Secreting Cells
2.2. Plpp5, Clptm1l and Itm2c Are Highly Conserved between Mice and Humans
2.3. Confirmation of Plpp5, Clptm1l and Itm2c Surface Expression and Prediction of Membrane Topology
2.4. PLPP5, CLPTM1L and ITM2C Are Highly Expressed in Multiple Myeloma
2.5. Generation of Plpp5, Clptm1l and Itm2c Knock-Out Mouse Models
2.6. Characterization of Plpp5, Clptm1l and Itm2c Deficient Mice
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Bioinformatic Analysis
4.3. Generation, Expression and Detection of Tagged Proteins
4.4. Patient Samples
4.5. Flow Cytometry
4.5.1. Mouse
4.5.2. Human
4.6. Multiple Myeloma Cell Lines
4.7. Immunohistochemistry
4.8. B Cell Isolation and Cell Culture
4.9. RNA Isolation and qRT-PCR
4.9.1. Mouse
4.9.2. Human
4.10. ELISA
4.11. Retroviral Transduction of B Cells
4.12. Immunization
4.13. ELISpot
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hiepe, F.; Radbruch, A. Plasma cells as an innovative target in autoimmune disease with renal manifestations. Nat. Rev. Nephrol. 2016, 12, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Kyle, R.A.; Rajkumar, S.V. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia 2009, 23, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Reff, M.; Carner, K.; Chambers, K.; Chinn, P.; Leonard, J.; Raab, R.; Newman, R.; Hanna, N.; Anderson, D. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood 1994, 83, 435–445. [Google Scholar] [PubMed]
- DiLillo, D.J.; Hamaguchi, Y.; Ueda, Y.; Yang, K.; Uchida, J.; Haas, K.M.; Kelsoe, G.; Tedder, T.F. Maintenance of long-lived plasma cells and serological memory despite mature and memory B cell depletion during CD20 immunotherapy in mice. J. Immunol. 2008, 180, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Hsi, E.D.; Steinle, R.; Balasa, B.; Szmania, S.; Draksharapu, A.; Shum, B.P.; Huseni, M.; Powers, D.; Nanisetti, A.; Zhang, Y.; et al. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin. Can. Res. 2008, 14, 2775–2784. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.-T.; Dillon, M.; Song, W.; Leiba, M.; Li, X.-F.; Burger, P.; Lee, A.I.; Podar, K.; Hideshima, T.; Rice, A.G.; et al. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu. Blood 2008, 112, 1329–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Weers, M.; Tai, Y.-T.; van der Veer, M.S.; Bakker, J.M.; Vink, T.; Jacobs, D.C.H.; Oomen, L.A.; Peipp, M.; Valerius, T.; Slootstra, J.W.; et al. Daratumumab, a novel therapeutic human CD38 monoclonal antibody, induces killing of multiple myeloma and other hematological tumors. J. Immunol. 2011, 186, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Liao, Y.; Willis, S.N.; Taubenheim, N.; Inouye, M.; Tarlinton, D.M.; Smyth, G.K.; Hodgkin, P.D.; Nutt, S.L.; Corcoran, L.M. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 2015, 16, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Immgen Consortium. Available online: www.Immgen.Org (accessed on 9 May 2018).
- BLUEPRINT Consortium. Available online: www.blueprint-epigenome.eu/ (accessed on 18 July 2018).
- Kahsay, R.Y.; Gao, G.; Liao, L. An improved hidden markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 2005, 21, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Viklund, H.; Bernsel, A.; Skwark, M.; Elofsson, A. Spoctopus: A combined predictor of signal peptides and membrane protein topology. Bioinformatics 2008, 24, 2928–2929. [Google Scholar] [CrossRef] [PubMed]
- Wickham, L.; Benjannet, S.; Marcinkiewicz, E.; Chretien, M.; Seidah, N.G. Beta-amyloid protein converting enzyme 1 and brain-specific type ii membrane protein BRI3: Binding partners processed by furin. J. Neurochem. 2005, 92, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Puskas, L.G.; Man, I.; Szebeni, G.; Tiszlavicz, L.; Tsai, S.; James, M.A. Novel anti-CRR9/CLPTM1l antibodies with antitumorigenic activity inhibit cell surface accumulation, PI3K interaction, and survival signaling. Mol. Cancer Ther. 2016. [Google Scholar] [CrossRef] [PubMed]
- Human Protein Atlas v18. Available online: https://www.Proteinatlas.Org/ensg00000049656-clptm1l/tissue/spleen#img (accessed on 8 May 2018).
- Kallies, A.; Hasbold, J.; Tarlinton, D.M.; Dietrich, W.; Corcoran, L.M.; Hodgkin, P.D.; Nutt, S.L. Plasma cell ontogeny defined by quantitative changes in Blimp-1 expression. J. Exp. Med. 2004, 200, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Anderson, K.C. Immune therapies in multiple myeloma. Clin. Cancer Res. 2016, 22, 5453–5460. [Google Scholar] [CrossRef] [PubMed]
- Sherbenou, D.W.; Behrens, C.R.; Su, Y.; Wolf, J.L.; Martin, T.G., 3rd; Liu, B. The development of potential antibody-based therapies for myeloma. Blood Rev. 2015, 29, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, M.; Harigai, M.; Momohara, S.; Ball, E.; Abe, J.; Furuichi, K.; Kamatani, N. Cloning and characterization of Dppl1 and Dppl2, representatives of a novel type of mammalian phosphatidate phosphatase. Gene 2007, 399, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Dong, Q.Z.; Ren, N.; Zhu, J.J.; Zhou, H.J.; Sun, H.J.; Wang, G.; Zhang, X.F.; Xue, Y.H.; Jia, H.L.; et al. Downregulation of HTPAP transcript variant 1 correlates with tumor metastasis and poor survival in patients with hepatocellular carcinoma. Cancer Sci. 2011, 102, 583–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard-Pierrot, I.; Gruel, N.; Stransky, N.; Vincent-Salomon, A.; Reyal, F.; Raynal, V.; Vallot, C.; Pierron, G.; Radvanyi, F.; Delattre, O. Characterization of the recurrent 8p11-12 amplicon identifies Ppapdc1b, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res. 2008, 68, 7165–7175. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, S.F.; Gruel, N.; Nicolle, R.; Chapeaublanc, E.; Delattre, O.; Radvanyi, F.; Bernard-Pierrot, I. Ppapdc1b and Whsc1l1 are common drivers of the 8p11-12 amplicon, not only in breast tumors but also in pancreatic adenocarcinomas and lung tumors. Am. J. Pathol. 2013, 183, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Engelhard, M. Anti-CD20 antibody treatment of non-hodgkin lymphomas. Clin. Immunol. 2016, 172, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Franks, S.E.; Getahun, A.; Hogarth, P.M.; Cambier, J.C. Targeting B cells in treatment of autoimmunity. Curr. Opin. Immunol. 2016, 43, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, Z.; Chen, Q.; Lai, Y.; Wang, Z.; Sun, L.; Luo, X.; Wang, X. Prognostic significance of CLPTM1L expression and its effects on migration and invasion of human lung cancer cells. Cancer Biomark. 2016, 16, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Bosley, A.D.; Thompson, A.; Hoskins, J.W.; Cheuk, A.; Collins, I.; Parikh, H.; Xiao, Z.; Ylaya, K.; Dzyadyk, M.; et al. CLPTM1L promotes growth and enhances aneuploidy in pancreatic cancer cells. Cancer Res. 2014, 74, 2785–2795. [Google Scholar] [CrossRef] [PubMed]
- James, M.A.; Wen, W.; Wang, Y.; Byers, L.A.; Heymach, J.V.; Coombes, K.R.; Girard, L.; Minna, J.; You, M. Functional characterization of CLPTM1L as a lung cancer risk candidate gene in the 5p15.33 locus. PLoS ONE 2012, 7, e36116. [Google Scholar] [CrossRef] [PubMed]
- Wauters, E.; Smeets, D.; Coolen, J.; Verschakelen, J.; De Leyn, P.; Decramer, M.; Vansteenkiste, J.; Janssens, W.; Lambrechts, D. The TERT-CLPTM1L locus for lung cancer predisposes to bronchial obstruction and emphysema. Eur. Respir. J. 2011, 38, 924–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, G.M.; Amundadottir, L.; Fuchs, C.S.; Kraft, P.; Stolzenberg-Solomon, R.Z.; Jacobs, K.B.; Arslan, A.A.; Bueno-de-Mesquita, H.B.; Gallinger, S.; Gross, M.; et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010, 42, 224–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, U.; Hutter, C.M.; Hsu, L.; Schumacher, F.R.; Conti, D.V.; Carlson, C.S.; Edlund, C.K.; Haile, R.W.; Gallinger, S.; Zanke, B.W.; et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum. Genet. 2012, 131, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, G.; Zhao, Y.; Song, X.; Chen, H.; Mao, Y.; Lu, D. Fine-mapping of a region of chromosome 5p15.33 (TERT-CLPTM1L) suggests a novel locus in TERT and a CLPTM1L haplotype are associated with glioma susceptibility in a chinese population. Int. J. Cancer 2012, 131, 1569–1576. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, C.; Rapley, E.A.; Seal, S.; Pernet, D.; Renwick, A.; Hughes, D.; Ricketts, M.; Linger, R.; Nsengimana, J.; Deloukas, P.; et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat. Genet. 2010, 42, 604–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, Z.; Tao, K.; Chen, G.; Chen, Q.; Tang, J.; Luo, X.; Yin, P.; Tang, J.; Wang, X. CLPTM1L is overexpressed in lung cancer and associated with apoptosis. PLoS ONE 2012, 7, e52598. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Fluhrer, R.; Haass, C. Substrate requirements for SPPL2B-dependent regulated intramembrane proteolysis. J. Biol. Chem. 2009, 284, 5662–5670. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.; Rebelo, S.; Santos, M.; Cotrim, C.Z.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A. BRI2 and BRI3 are functionally distinct phosphoproteins. Cell. Signal. 2016, 28, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, G.; Li, C.; Zhao, S. BRI3, a novel gene, participates in tumor necrosis factor-alpha-induced cell death. Biochem. Biophys. Res. Commun. 2003, 311, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.C.; Kim, J.; Kim, T.H.; Cho, S.Y.; Park, S.S.; Kim, K.D.; Lee, S.H. Cloning and characterization of a type II integral transmembrane protein gene, Itm2c, that is highly expressed in the mouse brain. Mol. Cells 2001, 12, 391–397. [Google Scholar] [PubMed]
- Vidal, R.; Calero, M.; Revesz, T.; Plant, G.; Ghiso, J.; Frangione, B. Sequence, genomic structure and tissue expression of human BRI3, a member of the Bri gene family. Gene 2001, 266, 95–102. [Google Scholar] [CrossRef]
- Matsuda, S.; Matsuda, Y.; D’Adamio, L. BRI3 inhibits amyloid precursor protein processing in a mechanistically distinct manner from its homologue dementia gene BRI2. J. Biol. Chem. 2009, 284, 15815–15825. [Google Scholar] [CrossRef] [PubMed]
- Rengaraj, D.; Gao, F.; Liang, X.H.; Yang, Z.M. Expression and regulation of type II integral membrane protein family members in mouse male reproductive tissues. Endocrine 2007, 31, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Farley, F.W.; Soriano, P.; Steffen, L.S.; Dymecki, S.M. Widespread recombinase expression using Flper (Flipper) mice. Genesis 2000, 28, 106–110. [Google Scholar] [CrossRef]
- Schwenk, F.; Baron, U.; Rajewsky, K. A cre-transgenic mouse strain for the ubiquitous deletion of loxp-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995, 23, 5080–5081. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. Emboss: The European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Kalff, A.; Chow, A.; Khong, T.; Spencer, A. Dysregulated class I histone deacetylases are indicators of poor prognosis in multiple myeloma. Epigenetics 2014, 9, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed]








| Genotype: | Clptm1l+/+ | Clptm1ldel/+ | Clptm1ldel/del |
|---|---|---|---|
| Expected | 13.25 | 26.5 | 13.25 |
| Observed | 21 | 29 | 3 * |
| Total Number Born | Number Survived Past Day 2 |
|---|---|
| 37 | 5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trezise, S.; Karnowski, A.; Fedele, P.L.; Mithraprabhu, S.; Liao, Y.; D’Costa, K.; Kueh, A.J.; Hardy, M.P.; Owczarek, C.M.; Herold, M.J.; et al. Mining the Plasma Cell Transcriptome for Novel Cell Surface Proteins. Int. J. Mol. Sci. 2018, 19, 2161. https://doi.org/10.3390/ijms19082161
Trezise S, Karnowski A, Fedele PL, Mithraprabhu S, Liao Y, D’Costa K, Kueh AJ, Hardy MP, Owczarek CM, Herold MJ, et al. Mining the Plasma Cell Transcriptome for Novel Cell Surface Proteins. International Journal of Molecular Sciences. 2018; 19(8):2161. https://doi.org/10.3390/ijms19082161
Chicago/Turabian StyleTrezise, Stephanie, Alexander Karnowski, Pasquale L. Fedele, Sridurga Mithraprabhu, Yang Liao, Kathy D’Costa, Andrew J. Kueh, Matthew P. Hardy, Catherine M. Owczarek, Marco J. Herold, and et al. 2018. "Mining the Plasma Cell Transcriptome for Novel Cell Surface Proteins" International Journal of Molecular Sciences 19, no. 8: 2161. https://doi.org/10.3390/ijms19082161