The Role of Epigenetic Regulation in Epstein-Barr Virus-Associated Gastric Cancer
Abstract
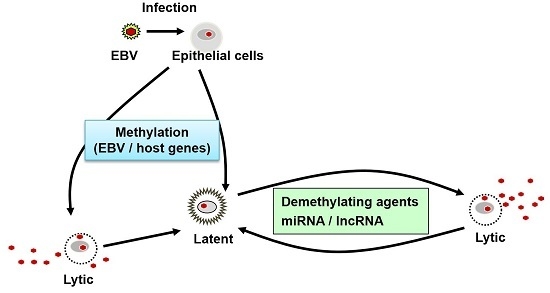
:1. Introduction
2. The Epigenetic Regulation of Latent Epstein–Barr Virus (EBV) Gene Expression
3. DNA Hypermethylation in the Host Genome
4. MicroRNA (miRNA)
4.1. EBV Encoded miRNAs
4.2. Host miRNAs
5. Long Non-Coding RNA (lncRNA)
5.1. EBV Encoded lncRNAs
5.2. Host lncRNAs
6. Demethylating Agents in EBV-Associated Gastric Cancer
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Bollschweiler, E.; Berlth, F.; Baltin, C.; Mönig, S.; Hölscher, A.H. Treatment of early gastric cancer in the Western World. World J. Gastroenterol. 2014, 20, 5672–5678. [Google Scholar] [CrossRef] [PubMed]
- Piazuelo, M.B.; Epplein, M.; Correa, P. Gastric cancer: An infectious disease. Infect. Dis. Clin. N. Am. 2010, 24, 853–869. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H.; Schulte-Holthausen, H.; Klein, G.; Henle, W.; Henle, G.; Clifford, P.; Santesson, L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 1970, 228, 1056–1058. [Google Scholar] [CrossRef] [PubMed]
- Burke, A.P.; Yen, T.S.; Shekitka, K.M.; Sobin, L.H. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod. Pathol. 1990, 3, 377–380. [Google Scholar] [PubMed]
- Takada, K. Epstein-Barr virus and gastric carcinoma. Mol. Pathol. 2000, 53, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Shibata, D.; Weiss, L.M. Epstein-Barr virus-associated gastric adenocarcinoma. Am. J. Pathol. 1992, 140, 769–774. [Google Scholar] [PubMed]
- Tokunaga, M.; Land, C.E.; Uemura, Y.; Tokudome, T.; Tanaka, S.; Sato, E. Epstein-Barr virus in gastric carcinoma. Am. J. Pathol. 1993, 143, 1250–1254. [Google Scholar] [PubMed]
- Imai, S.; Koizumi, S.; Sugiura, M.; Tokunaga, M.; Uemura, Y.; Yamamoto, N.; Tanaka, S.; Sato, E.; Osato, T. Gastric carcinoma: Monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc. Natl. Acad. Sci. USA 1994, 91, 9131–9135. [Google Scholar] [CrossRef] [PubMed]
- Fukayama, M.; Hayashi, Y.; Iwasaki, Y.; Chong, J.; Ooba, T.; Takizawa, T.; Koike, M.; Mizutani, S.; Miyaki, M.; Hirai, K. Epstein-Barr virus-associated gastric carcinoma and Epstein-Barr virus infection of the stomach. Lab. Investig. 1994, 71, 73–81. [Google Scholar] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [Green Version]
- Kawazoe, A.; Kuwata, T.; Kuboki, Y.; Shitara, K.; Nagatsuma, A.K.; Aizawa, M.; Yoshino, T.; Doi, T.; Ohtsu, A.; Ochiai, A. Clinicopathological features of programmed death ligand 1 expression with tumor-infiltrating lymphocyte, mismatch repair, and Epstein-Barr virus status in a large cohort of gastric cancer patients. Gastric Cancer 2017, 20, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Derks, S.; Liao, X.; Chiaravalli, A.M.; Xu, X.; Camargo, M.C.; Solcia, E.; Sessa, F.; Fleitas, T.; Freeman, G.J.; Rodig, S.J.; et al. Abundant PD-L1 expression in Epstein-Barr Virus-infected gastric cancers. Oncotarget 2016, 7, 32925–32932. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Patel, K.; Singhi, A.D.; Ren, B.; Zhu, B.; Shaikh, F.; Sun, W. Programmed death-ligand 1 expression is common in gastric cancer associated with Epstein-Barr virus or microsatellite instability. Am. J. Surg. Pathol. 2016, 40, 1496–1506. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.S.; Uozaki, H.; Chong, J.M.; Ushiku, T.; Sakuma, K.; Ishikawa, S.; Hino, R.; Barua, R.R.; Iwasaki, Y.; Arai, K.; et al. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin. Cancer Res. 2006, 12, 2995–3002. [Google Scholar] [PubMed]
- Kusano, M.; Toyota, M.; Suzuki, H.; Akino, K.; Aoki, F.; Fujita, M.; Hosokawa, M.; Shinomura, Y.; Imai, K.; Tokino, T. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer 2006, 106, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, A.; Matsusaka, K.; Aburatani, H.; Fukayama, M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012, 72, 3445–3450. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Nakamura, M.; Nishikawa, J.; Sakai, K.; Zhang, Y.; Saito, M.; Morishige, A.; Oga, A.; Sasaki, K.; Suehiro, Y.; et al. Identification of genes specifically methylated in Epstein-Barr virus-associated gastric carcinomas. Cancer Sci. 2013, 104, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Nishikawa, J.; Okada, T.; Morishige, A.; Sakai, K.; Nakamura, M.; Kiyotoki, S.; Hamabe, K.; Okamoto, T.; Oga, A.; et al. Role of DNA methylation in the development of Epstein-Barr virus-associated gastric carcinoma. J. Med. Virol. 2013, 85, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Niller, H.H.; Wolf, H.; Minarovits, J. Epigenetic dysregulation of the host cell genome in Epstein-Barr virus-associated neoplasia. Semin. Cancer Biol. 2009, 19, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Ernberg, I.; Falk, K.; Minarovits, J.; Busson, P.; Tursz, T.; Masucci, M.G.; Klein, G. The role of methylation in the phenotype-dependent modulation of Epstein-Barr nuclear antigen 2 and latent membrane protein genes in cells latently infected with Epstein-Barr virus. J. Gen. Virol. 1989, 70, 2989–3002. [Google Scholar] [CrossRef] [PubMed]
- Masucci, M.G.; Contreras-Salazar, B.; Ragnar, E.; Falk, K.; Minarovits, J.; Ernberg, I.; Klein, G. 5-Azacytidine up regulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt’s lymphoma line rael. J. Virol. 1989, 63, 3135–3141. [Google Scholar] [PubMed]
- Sugiura, M.; Imai, S.; Tokunaga, M.; Koizumi, S.; Uchizawa, M.; Okamoto, K.; Osato, T. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: Unique viral latency in the tumour cells. Br. J. Cancer 1996, 74, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Wang, Y.; Wang, X.F.; Liang, H.; Yan, L.P.; Huang, B.H.; Zhao, P. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J. Gastroenterol. 2005, 11, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Morgan, D.R.; Meyers, M.O.; Dominguez, R.L.; Martinez, E.; Kakudo, K.; Kuan, P.F.; Banet, N.; Muallem, H.; Woodward, K.; et al. Epstein-barr virus infected gastric adenocarcinoma expresses latent and lytic viral transcripts and has a distinct human gene expression profile. Infect. Agent Cancer 2012, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Strong, M.J.; Laskow, T.; Nakhoul, H.; Blanchard, E.; Liu, Y.; Wang, X.; Baddoo, M.; Lin, Z.; Yin, Q.; Flemington, E.K. Latent expression of the Epstein-Barr virus (EBV)-encoded major histocompatibility complex class I TAP inhibitor, BNLF2a, in EBV-positive gastric carcinomas. J. Virol. 2015, 89, 10110–10114. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.; Oliveira, C.; Malta, M.; Sousa, H. Epstein-Barr virus gene expression and latency pattern in gastric carcinomas: A systematic review. Future Oncol. 2017, 13, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.F.; Minarovits, J.; Cao, S.L.; Contreras-Salazar, B.; Rymo, L.; Falk, K.; Klein, G.; Ernberg, I. Variable expression of latent membrane protein in nasopharyngeal carcinoma can be related to methylation status of the Epstein-Barr virus BNLF-1 5’-flanking region. J. Virol. 1991, 65, 1558–1567. [Google Scholar] [PubMed]
- Iizasa, H.; Nanbo, A.; Nishikawa, J.; Jinushi, M.; Yoshiyama, H. Epstein-Barr Virus (EBV)-associated gastric carcinoma. Viruses 2012, 4, 3420–3439. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, J.; Yoshiyama, H.; Iizasa, H.; Kanehiro, Y.; Nakamura, M.; Nishimura, J.; Saito, M.; Okamoto, T.; Sakai, K.; Suehiro, Y.; et al. Epstein-barr virus in gastric carcinoma. Cancers 2014, 6, 2259–2274. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.H.; Lee, S.; Kim, W.H.; Lee, H.W.; Kim, J.C.; Rhyu, M.G.; Ro, J.Y. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am. J. Pathol. 2002, 160, 787–794. [Google Scholar] [CrossRef]
- Vo, Q.N.; Geradts, J.; Gulley, M.L.; Boudreau, D.A.; Bravo, J.C.; Schneider, B.G. Epstein-Barr virus in gastric adenocarcinomas: Association with ethnicity and CDKN2A promoter methylation. J. Clin. Pathol. 2002, 55, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.M.; Sakuma, K.; Sudo, M.; Ushiku, T.; Uozaki, H.; Shibahara, J.; Nagai, H.; Funata, N.; Taniguchi, H.; Aburatani, H.; et al. Global and non-random CpG-island methylation in gastric carcinoma associated with Epstein-Barr virus. Cancer Sci. 2003, 94, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.M.; Lee, H.G.; Cho, S.G.; Kwon, S.H.; Yoon, H.; Kwon, H.J.; Lee, J.H.; Kim, H.; Park, P.G.; Kim, H.; et al. Hypermethylation of the interferon regulatory factor 5 promoter in Epstein-Barr virus-associated gastric carcinoma. J. Microbiol. 2015, 53, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Liang, Q.; Wang, J.; Wang, K.; Gao, J.; Zhang, J.; Zeng, Y.; Chiu, P.W.; Ng, E.K.; Sung, J.J. REC8 functions as a tumor suppressor and is epigenetically downregulated in gastric cancer, especially in EBV-positive subtype. Oncogene 2017, 36, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, J.H.; Baek, S.J.; Kim, S.Y.; Kim, Y.S. Specific expression and methylation of SLIT1, SLIT2, SLIT3, and miR-218 in gastric cancer subtypes. Int. J. Oncol. 2016, 48, 2497–2507. [Google Scholar] [CrossRef] [PubMed]
- Hino, R.; Uozaki, H.; Murakami, N.; Ushiku, T.; Shinozaki, A.; Ishikawa, S.; Morikawa, T.; Nakaya, T.; Sakatani, T.; Takada, K.; et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009, 69, 2766–2774. [Google Scholar] [CrossRef] [PubMed]
- Lennette, E.T.; Winberg, G.; Yadav, M.; Enblad, G.; Klein, G. Antibodies to LMP2A/2B in EBV-carrying malignancies. Eur. J. Cancer 1995, 31A, 1875–1878. [Google Scholar] [CrossRef]
- Tsai, C.L.; Li, H.P.; Lu, Y.J.; Hsueh, C.; Liang, Y.; Chen, C.L.; Tsao, S.W.; Tse, K.P.; Yu, J.S.; Chang, Y.S. Activation of DNA methyltransferase1 by EBV LMP1 Involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 2006, 66, 11668–11676. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.N.; Tsai, C.L.; Tse, K.P.; Chang, H.Y.; Chang, Y.S. The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc. Natl. Acad. Sci. USA 2002, 99, 10084–10089. [Google Scholar] [CrossRef] [PubMed]
- Kida, Y.; Miyauchi, K.; Takano, Y. Gastric adenocarcinoma with differentiation to sarcomatous components associated with monoclonal Epstein-Barr virus infection and LMP-1 expression. Virchows Arch. A Pathol. Anat. Histopathol. 1993, 423, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Matsusaka, K.; Kaneda, A.; Nagae, G.; Ushiku, T.; Kikuchi, Y.; Hino, R.; Uozaki, H.; Seto, Y.; Takada, K.; Aburatani, H.; et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011, 71, 7187–7197. [Google Scholar] [CrossRef] [PubMed]
- Matsusaka, K.; Funata, S.; Fukuyo, M.; Seto, Y.; Aburatani, H.; Fukayama, M.; Kaneda, A. Epstein-Barr virus infection induces genome-wide de novo DNA methylation in non-neoplastic gastric epithelial cells. J. Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Fan, J.; Belasco, J.G. MicroRNA direct rapid MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 4034–4039. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, P.; Maragkakis, M.; Papadopoulos, G.L.; Reczko, M.; Hatzigeorgiou, A.G. Lost in translation: An assessment and perspective for computational microRNA target identification. Bioinformatics 2009, 25, 3049–3055. [Google Scholar]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; John, B.; Enright, A.J.; Sander, C.; Tuschl, T. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Schater, A.; Lu, S.; Bilello, J.P.; Desrosiers, R.C.; Edwards, R.; Raab-Traub, N.; Cullen, B.R. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006, 2, e23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, J.; Cosmopoulos, K.; Pegtel, M.; Hopmans, E.; Murray, P.; Middeldorp, J.; Shapiro, M.; Thorley-Lawson, D.A. A novel persistence associated EBV miRNA expression profile is distributed in neoplasia. PLoS Pathog. 2011, 7, e1002193. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Choi, H.; Lee, S.K. Epstein-Barr virus miR-BART20–5p regulates cell proliferation and apoptosis by targeting BAD. Cancer Lett. 2015, 356, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki-Ushiku, A.; Kunita, A.; Isogai, M.; Hibiya, T.; Ushiku, T.; Takada, K.; Fukayama, M. Profiling of Virus-Encoded MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma and Their Roles in Gastric Carcinogenesis. J. Virol. 2015, 89, 5581–5591. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Skalsky, R.L.; Cullen, B.R. EBV BART microRNAs target multiple pro-apoptotic cellular genes to promote epithelial cell survival. PLoS Pathog. 2015, 11, e1004979. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Miyata, M.; Kano, M.; Kondo, S.; Yoshizaki, T.; Iizasa, H. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J. Virol. 2015, 89, 2684–2697. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, X.; Zeng, Z.; Li, Q.; Gong, Z.; Liao, Q.; Li, X.; Chen, P.; Xiang, B.; Zhang, W.; et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget 2016, 7, 36783–36799. [Google Scholar] [CrossRef] [PubMed]
- Iizasa, H.; Wulff, B.E.; Alla, N.R.; Maragkakis, M.; Megraw, M.; Hatzigeorgiou, A.; Iwakiri, D.; Takada, K.; Wiedmer, A.; Showe, L.; et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 2010, 285, 33358–33370. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Choi, H.; Kim, H.; Lee, S.K. MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J. Virol. 2014, 88, 9027–9037. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Tsai, M.H.; Shumilov, A.; Poirey, R.; Bannert, H.; Middeldorp, J.M.; Feederle, R.; Delecluse, H.J. The Epstein-Barr virus BART miRNA cluster of the M81 strain modulates multiple functions in primary B cells. PLoS Pathog. 2015, 11, e1005344. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Chen, G.H.; Chen, Y.H.; Liu, C.Y.; Chang, K.P.; Chang, Y.S.; Chen, H.C. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS ONE 2010, 5, e12745. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Huang, J.; Wu, F.Y.; Liao, G.; Hutt-Fletcher, L.; Hayward, S.D. Regulation of expression of the Epstein-Barr virus BamHI-A rightward transcripts. J. Virol. 2005, 79, 1724–1733. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.N.; Song, Y.J.; Lee, S.K. The role of promoter methylation in Epstein-Barr virus (EBV) microRNA expression in EBV-infected B cell lines. Exp. Mol. Med. 2011, 43, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, A.; Sakatani, T.; Ushiku, T.; Hino, R.; Isogai, M.; Ishikawa, S.; Uozaki, H.; Takada, K.; Fukayama, M. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 2010, 70, 4719–4727. [Google Scholar] [CrossRef] [PubMed]
- Marquitz, A.R.; Mathur, A.; Chugh, P.E.; Dittmer, D.P.; Raab-Traub, N. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J. Virol. 2014, 88, 1389–1393. [Google Scholar] [CrossRef] [PubMed]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. MicroRNA targeting dicer for metastasis control. Cell 2010, 141, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Merritt, W.M.; Lin, Y.G.; Han, L.Y.; Kamat, A.A.; Spannuth, W.A.; Schmandt, R.; Urbauer, D.; Pennacchio, L.A.; Cheng, J.F.; Nick, A.M.; et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008, 359, 2641–2650. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Marquitz, A.R.; Mathur, A.; Edwards, R.H.; Raab-Traub, N. Host Gene Expression Is Regulated by Two Types of Noncoding RNAs Transcribed from the Epstein-Barr Virus BamHI A Rightward Transcript Region. J. Virol. 2015, 89, 11256–11268. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Nagano, O.; Yae, T.; Tamada, M.; Motohara, T.; Oshima, H.; Oshima, M.; Ikeda, T.; Asaba, R.; Yagi, H.; et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc- and thereby promotes tumor growth. Cancer Cell 2011, 19, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Conforti, F.; Yang, A.L.; Piro, M.C.; Mellone, M.; Terrinoni, A.; Candi, E.; Tucci, P.; Thomas, G.J.; Knight, R.A.; Melino, G.; et al. PIR2/Rnf144B regulates epithelial homeostasis by mediating degradation of p21WAF1 and p63. Oncogene 2013, 32, 4758–4765. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Moss, W.; O’Grady, T.; Concha, M.; Strong, M.J.; Wang, X.; Yu, Y.; Baddoo, M.; Zhang, K.; Fewell, C.; et al. New noncoding lytic transcripts derived from the Epstein-Barr Virus Latency Origin of Replication, oriP, are hyperedited, bind the paraspeckle protein, NONO/p54nrb, and support viral lytic transcription. J. Virol. 2015, 89, 7120–7132. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Ji, Y.; Hu, D.; Chen, B.; Zhang, H.; Li, C.; Chen, G.; Luo, X.; Zheng, X.W.; Lin, X. SNHG8 is identified as a key regulator of epstein-barr virus (EBV)-associated gastric cancer by an integrative analysis of lncRNA and mRNA expression. Oncotarget 2016, 7, 80990–81002. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Bi, J.; Xue, X.; Zheng, L.; Zhi, K.; Hua, J.; Fang, G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012, 279, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Vella, S.; Miele, M.; Timoneri, F.; Di Bella, M.; Bosi, S.; Sciveres, M.; Conaldi, P.G. Global profiling of viral and cellular non-coding RNAs in Epstein-Barr virus-induced lymphoblastoid cell lines and released exosome cargos. Cancer Lett. 2017, 388, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Piedade, D.; Azevedo-Pereira, J.M. The role of microRNAs in the pathogenesis of herpesvirus infection. Viruses 2016, 8, 156. [Google Scholar] [CrossRef]
- Giudice, A.; D’Arena, G.; Crispo, A.; Tecce, M.F.; Nocerino, F.; Grimaldi, M.; Rotondo, E.; D’Ursi, A.M.; Scrima, M.; Galdiero, M.; et al. Role of viral miRNAs and epigenetic modifications in Epstein-Barr virus-associated gastric carcinogenesis. Oxid. Med. Cell. Longev. 2016, 2016, 6021934. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Meltzer, S.J. Gastric cancer in the era of precision medicine. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Silverman, L.R.; Demakos, E.P.; Peterson, B.L.; Kornblith, A.B.; Holland, J.C.; Odchimar-Reissig, R.; Stone, R.M.; Nelson, D.; Powell, B.L.; DeCastro, C.M.; et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: A study of the cancer and leukemia group B. J. Clin. Oncol. 2002, 20, 2429–2440. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.; Issa, J.P.; Rosenfeld, C.S.; Bennett, J.M.; Albitar, M.; DiPersio, J.; Klimek, V.; Slack, J.; de Castro, C.; Ravandi, F.; et al. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer 2006, 106, 1794–1803. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef]
- Benton, C.B.; Thomas, D.A.; Yang, H.; Ravandi, F.; Rytting, M.; O’Brien, S.; Franklin, A.R.; Borthakur, G.; Dara, S.; Kwari, M.; et al. Safety and clinical activity of 5-aza-2′-deoxycytidine (decitabine) with or without Hyper-CVAD in relapsed/refractory acute lymphocytic leukaemia. Br. J. Haematol. 2014, 167, 356–365. [Google Scholar] [CrossRef]
- Mayer, J.; Arthur, C.; Delaunay, J.; Mazur, G.; Thomas, X.G.; Wierzbowska, A.; Ravandi, F.; Berrak, E.; Jones, M.; Li, Y.; et al. Multivariate and subgroup analyses of a randomized, multinational, phase 3 trial of decitabine vs treatment choice of supportive care or cytarabine in older patients with newly diagnosed acute myeloid leukemia and poor- or intermediate-risk cytogenetics. BMC Cancer 2014, 14, 69. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Nishikawa, J.; Saito, M.; Sakai, K.; Sasaki, S.; Hashimoto, S.; Okamoto, T.; Suehiro, Y.; Yamasaki, T.; Sakaida, I. Decitabine inhibits tumor cell proliferation and up-regulates e-cadherin expression in Epstein-Barr virus-associated gastric cancer. J. Med. Virol. 2017, 89, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.P.; Rechsteiner, M.P.; Berger, C.; Sigrist, J.A.; Nadal, D.; Bernasconi, M. Zebularine reactivates silenced E-cadherin but unlike 5-Azacytidine does not induce switching from latent to lytic Epstein-Barr virus infection in Burkitt’s lymphoma Akata cells. Mol. Cancer 2007, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Gradoville, L.; Daigle, D.; Miller, G. De novo protein synthesis is required for lytic cycle reactivation of Epstein-Barr virus, but not Kaposi’s sarcoma-associated herpesvirus, in response to histone deacetylase inhibitors and protein kinase C agonists. J. Virol. 2007, 81, 9279–9291. [Google Scholar] [CrossRef] [PubMed]
- Countryman, J.K.; Gradoville, L.; Miller, G. Histone hyperacetylation occurs on promoters of lytic cycle regulatory genes in Epstein-Barr virus-infected cell lines which are refractory to disruption of latency by histone deacetylase inhibitors. J. Virol. 2008, 82, 4706–4719. [Google Scholar] [CrossRef] [PubMed]
- Countryman, J.; Gradoville, L.; Bhaduri-McIntosh, S.; Ye, J.; Heston, L.; Himmelfarb, S.; Shedd, D.; Miller, G. Stimulus duration and response time independently influence the kinetics of lytic cycle reactivation of Epstein-Barr virus. J. Virol. 2009, 83, 10694–10709. [Google Scholar] [CrossRef] [PubMed]
- Larochelle, B.; Flamand, L.; Gourde, P.; Beauchamp, D.; Gosselin, J. Epstein-Barr virus infects and induces apoptosis in human neutrophils. Blood 1998, 92, 291–299. [Google Scholar] [PubMed]
- Inman, G.J.; Binné, U.K.; Parker, G.A.; Farrell, P.J.; Allday, M.J. Activators of the Epstein-Barr virus lytic program concomitantly induce apoptosis, but lytic gene expression protects from cell death. J. Virol. 2001, 75, 2400–2410. [Google Scholar] [CrossRef] [PubMed]
- Isobe, Y.; Sugimoto, K.; Yang, L.; Tamayose, K.; Egashira, M.; Kaneko, T.; Takada, K.; Oshimi, K. Epstein-Barr virus infection of human natural killer cell lines and peripheral blood natural killer cells. Cancer Res. 2004, 64, 2167–2174. [Google Scholar] [CrossRef] [PubMed]
- Oussaief, L.; Hippocrate, A.; Clybouw, C.; Rampanou, A.; Ramirez, V.; Desgranges, C.; Vazquez, A.; Khelifa, R.; Joab, I. Activation of the lytic program of the Epstein-Barr virus in Burkitt's lymphoma cells leads to a two steps downregulation of expression of the proapoptotic protein BimEL, one of which is EBV-late-gene expression dependent. Virology 2009, 387, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Zuo, Q.; Pilrose, J.; Wang, Y.; Shen, C.; Li, M.; Wulfridge, P.; Matei, D.; Nephew, K.P. Decitabine reactivated pathways in platinum resistant ovarian cancer. Oncotarget 2014, 5, 3579–3589. [Google Scholar] [CrossRef] [PubMed]
- Fukayama, F.; Ushiku, T. Epstein-Barr virus-associated gastric carcinoma. Pathol. Res. Pract. 2011, 207, 529–537. [Google Scholar] [CrossRef] [PubMed]




| Type of Latency | Latent Gene Expression | Promoter Hypermethylation | EBV-Associated Diseases | ||
|---|---|---|---|---|---|
| Cp/Wp | Qp | LMP1p | |||
| Type I | EBNA1 | + | − | + | Burkitt lymphoma; Gastric cancer |
| Type II | EBNA1, LMP1, LMP2 | + | − | +/− | Nasopharyngeal cancer, Hodgikin disease, NK/T cell lymphoma |
| Type III | EBNA1–6, LMP1, LMP2 | − | − | − | Immunosuppressive lymphoma |
| miRNA | Target | Role of Targeted Gene | Reference | |
|---|---|---|---|---|
| EBV miRNAs | miR-BART3 | DICE1 | Apoptosis | [51] |
| FEM1B | Apoptosis | [51] | ||
| CASZ1a | Apoptosis | [51] | ||
| miR-BART4-5p | BID | Apoptosis | [50] | |
| miR-BART6-5p | DICER1 | Lytic replication-related genes | [51,54] | |
| DICER1 | Repressor of EMT | [51,60] | ||
| OCT1 | Apoptosis | [46,51] | ||
| miR-BART8 | ARID2 | Tumor suppressor gene | [51] | |
| miR-BART11 | FOXP1 | Cellular differentiation factor | [53] | |
| miR-BART16 | CREBBP | Apoptosis | [51] | |
| SH2B3 | Apoptosis | [51] | ||
| TOMM22 | Mitochondrial transporter | [51] | ||
| miR-BART20-5p | BAD | Apoptosis | [49] | |
| BZLF1/BRLF1 | Lytic replication-related genes | [49] | ||
| miR-BART22 | PPP3R1 | Apoptosis | [51] | |
| PAK2 | Apoptosis | [51] | ||
| TP53INP1 | Apoptosis | [51] | ||
| NDRG1 | Cellular differentiation factor | [52] | ||
| Host miRNA (Reduction) | miR-200 | ZEB1, ZEB2 | Repressor of EMT | [61,62] |
| miR-143-3p | KLF4, ELK1 | Stem cell factor | [61,62] | |
| miR-146b | STAT3 | Inflammation (IL-6) | [61,62] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishikawa, J.; Iizasa, H.; Yoshiyama, H.; Nakamura, M.; Saito, M.; Sasaki, S.; Shimokuri, K.; Yanagihara, M.; Sakai, K.; Suehiro, Y.; et al. The Role of Epigenetic Regulation in Epstein-Barr Virus-Associated Gastric Cancer. Int. J. Mol. Sci. 2017, 18, 1606. https://doi.org/10.3390/ijms18081606
Nishikawa J, Iizasa H, Yoshiyama H, Nakamura M, Saito M, Sasaki S, Shimokuri K, Yanagihara M, Sakai K, Suehiro Y, et al. The Role of Epigenetic Regulation in Epstein-Barr Virus-Associated Gastric Cancer. International Journal of Molecular Sciences. 2017; 18(8):1606. https://doi.org/10.3390/ijms18081606
Chicago/Turabian StyleNishikawa, Jun, Hisashi Iizasa, Hironori Yoshiyama, Munetaka Nakamura, Mari Saito, Sho Sasaki, Kanami Shimokuri, Masashi Yanagihara, Kouhei Sakai, Yutaka Suehiro, and et al. 2017. "The Role of Epigenetic Regulation in Epstein-Barr Virus-Associated Gastric Cancer" International Journal of Molecular Sciences 18, no. 8: 1606. https://doi.org/10.3390/ijms18081606