Plant Lectins and Lectin Receptor-Like Kinases: How Do They Sense the Outside?
Abstract
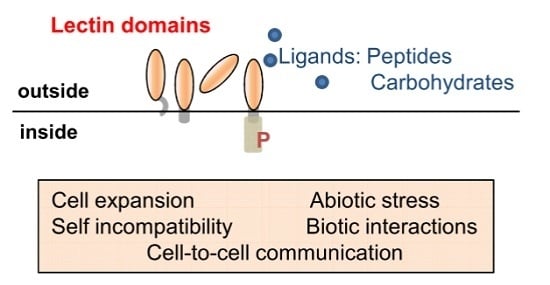
:1. Introduction
2. Occurrence of Lectin Domains in Plants
2.1. Distribution of Lectin Domains through Life Kingdoms
2.2. LecRKs, LecRLPs and LecPs Are Located at the Cell Surface and Exhibit PTMs: A Proteomics View
3. G-Type Lectin Receptor-Like Kinases
4. L-Type Lectin Receptor-Like Kinases
5. LysM Receptor-Like Kinases
6. Malectin Receptor-Like Kinases
7. Concluding Remarks
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Damme, E.; Lannoo, N.; Peumans, W. Plant lectins. Adv. Bot. Res. 2008, 48, 107–209. [Google Scholar] [CrossRef]
- Loris, R. Principles of structures of animal and plant lectins. Biochim. Biophys. Acta 2002, 1572, 198–208. [Google Scholar] [CrossRef]
- Carpita, N.; Gibeaut, D. Structural models of primary cell walls in flowering plants, consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Hematy, K.; Höfte, H. Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 2012, 63, 381–407. [Google Scholar] [CrossRef] [PubMed]
- Imberty, A.; Lortat-Jacob, H.; Pérez, S. Structural view of glucosaminoglycan-protein interactions. Carbohydr. Res. 2007, 342, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Gomord, V.; Fitchette, A.; Menu-Bouaouiche, L.; Saint-Jore-Dupas, C.; Plason, C.; Michaud, D.; Faye, L. Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 2010, 8, 564–587. [Google Scholar] [CrossRef] [PubMed]
- Aigal, S.; Claudinon, J.; Römer, W. Plasma membrane reorganization: A glycolipid gateway for microbes. Biochim. Biophys. Acta 2015, 1853, 858–871. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.; Men, S.; Grebe, M. Lipid function in plant cell polarity. Curr. Opin. Plant Biol. 2004, 7, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.; Bolam, D.; Gilbert, H.; Davies, G. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Drickamer, K. Convergent and divergent mechanisms of sugar recognition across kingdoms. Curr. Opin. Struct. Biol. 2014, 28, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.; Drickamer, K. Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 1996, 65, 441–473. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.; Singh, S.; Singh, S.; Rully, H.; Singh, L. Purification and characterization of a magnesium ion requiring N-acetyl-d-glucosamine specific lectin from seeds of Quercus ilex L. Biosci. Biotechnol. Biochem. 2011, 75, 1752–1757. [Google Scholar] [CrossRef] [PubMed]
- De Moura, K.; da Silva, H.; Dornelles, L.; Coelho, L.; Napoleão, T.; de Oliveira, M.; Paiva, P. Coagulant activity of water-soluble Moringa oleifera lectin is linked to lowering of electrical resistance and inhibited by monosaccharides and magnesium ions. Appl. Biochem. Biotechnol. 2016, 180, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Gupta, G.; Vijayan, M.; Surolia, A. Subunit assembly of plant lectins. Curr. Opin. Struct. Biol. 2007, 17, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Chrispeels, M.; Raikhel, N. Lectins, lectin genes, and their role in plant defense. Plant Cell 1991, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; van Damme, E. Nucleocytoplasmic plant lectins. Biochim. Biophys. Acta 2010, 1800, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Van Holle, S.; Smagghe, G.; van Damme, E. Overexpression of Nictaba-like lectin genes from Glycine max confers tolerance toward Pseudomonas syringae infection, aphid infestation and salt stress in transgenic Arabidopsis plants. Front. Plant Sci. 2016, 7, 1590. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Hervé, C.; Lescure, B.; Rougé, P. Lectin receptor kinases in plants. Crit. Rev. Plant Sci. 2002, 21, 379–399. [Google Scholar] [CrossRef]
- Hervé, C.; Dabos, P.; Galaud, J.; Rougé, P.; Lescure, B. Characterization of an Arabidopsis thaliana gene that defines a new class of putative plant receptor kinases with an extracellular lectin-like domain. J. Mol. Biol. 1996, 258, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Hervé, C.; Serres, J.; Dabos, P.; Canut, H.; Barre, A.; Rougé, P.; Lescure, B. Characterization of the Arabidopsis lecRK-a genes: Members of a superfamily encoding putative receptors with an extracellular domain homologous to legume lectins. Plant Mol. Biol. 1999, 39, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.; Knox, J.; Boraston, A. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 2013, 23, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, X.; Findley, S.; Wan, J.; Libault, M.; Nguyen, H.; Cannon, S.; Stacey, G. Molecular evolution of Lysin motif-type receptor-like kinases in plants. Plant Physiol. 2007, 144, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dernier, A.; Kessler, S.; Grossniklaus, U.T. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 2011, 62, 1581–1591. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.; Coggill, P.; Eberhardt, R.; Eddy, S.; Mistry, J.; Mitchell, A.; Potter, S.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, D.; Lim, Y.; Pai, H. The shooty callus induced by suppression of tobacco CHRK1 receptor-like kinase is a phenocopy of the tobacco genetic tumor. Plant Cell Rep. 2004, 23, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Takei, K.; Sakakibara, H.; Sun Cho, H.; Kim, D.; Kim, Y.; Min, S.; Kim, W.; Sohn, D.; Lim, Y.; et al. CHRK1, a chitinase-related receptor-like kinase, plays a role in plant development and cytokinin homeostasis in tobacco. Plant Mol. Biol. 2003, 53, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Labbé, J.; Muchero, W.; Yang, X.; Jawdy, S.; Kennedy, M.; Johnson, J.; Sreedasyam, A.; Schmutz, J.; Tuskan, G.; et al. Genome-wide analysis of lectin receptor-like kinases in Populus. BMC Genom. 2016, 17, 699. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Pandey, P.; Tuteja, N. Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol. Biol. 2012, 80, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Sharma, S.; Pandley, A.; Singh, K.; Upadhyay, S. Molecular characterization and global expression analysis of lectin receptor kinases in bread wheat (Triticum aestivum). PLoS ONE 2016, 11, e0153925. [Google Scholar]
- Hofberger, J.; Nsibo, D.; Govers, F.; Bouwmeester, K.; Schranz, M. A complex interplay of tandem- and whole-genome duplication drives expansion of the L-type lectin receptor kinase gene family in the Brassicaceae. Genome Biol. Evol. 2015, 7, 720–734. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, M.; Dwyer, K.; Hendershot, J.; Vrebalov, J.; Nasrallah, J.; Nasrallah, M. Self-incompatibility in the genus Arabidopsis. Characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 2001, 13, 527–643. [Google Scholar] [CrossRef]
- Jones, D.; Taylor, W.; Thornton, J. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Tordai, H.; Bányai, L.; Patthy, L. The PAN module: The N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999, 461, 63–67. [Google Scholar] [CrossRef]
- Shiu, S.; Bleecker, A. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed]
- Albenne, C.; Canut, H.; Jamet, E. Plant cell wall proteomics: The leadership of Arabidopsis thaliana. Front. Plant Sci. 2013, 4, 111. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-May, E.; Rose, J. Progress toward the tomato fruit cell wall proteome. Front. Plant Sci. 2013, 4, 159. [Google Scholar] [CrossRef] [PubMed]
- Ligat, L.; Lauber, E.; Albenne, C.; San Clemente, H.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Arlat, M.; Jamet, E. Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 2011, 11, 1798–1813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xin, W.; Wang, S.; Zhang, X.; Dai, H.; Sun, R.; Frazier, T.; Zhang, B.; Wang, Q. Xylem sap in cotton contains proteins that contribute to environmental stress response and cell wall development. Funct. Integr. Genom. 2015, 15, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Catalá, C.; Howe, K.; Hucko, S.; Rose, J.; Thannhauser, T. Towards characterization of the glycoproteome of tomato (Solanum lycopersicum) fruit using Concanavalin A lectin affinity chromatography and LC-MALDI-MS/MS analysis. Proteomics 2011, 11, 1530–1544. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Jamet, E.; Negroni, L.; der Garabedian, P.A.; Zivy, M.; Jouanin, L. A sub-proteome of Arabidopsis thaliana trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 2007, 58, 2503–2512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Giboulot, A.; Zivy, M.; Valot, B.; Jamet, E.; Albenne, C. Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 2011, 72, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Florance, H.; Johns, A.; Smirnoff, N. Ascorbate deficiency influences the leaf cell wall glycoproteome in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Medzihradszky, K.; Wang, Z.; Burlingame, A.; Chalkley, R. N-Glycopeptide profiling in Arabidopsis inflorescence. Mol. Cell. Proteom. 2016, 15, 2048–2054. [Google Scholar] [CrossRef] [PubMed]
- Borner, G.H.; Lilley, K.S.; Stevens, T.J.; Dupree, P. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 2003, 132, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Elortza, F.; Nühse, T.; Foster, L.; Stensballe, A.; Peck, S.; Jensen, O. Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteom. 2003, 2, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Elortza, F.; Shabaz, M.; Bunkenborg, J.; Foster, L.; Nühse, T.; Brodbeck, U.; Peck, S.; Jensen, O. Modification-specific proteomics of plasma membrane proteins: Identification and characterization of glycosylphosphatidylinositol-anchored proteins released upon phospholipase D treatment. J. Proteome Res. 2005, 5, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Kawamura, Y.; Uemura, M. Cold acclimation is accompanied by complex responses of glycosylphosphatidylinositol (GPI)-anchored proteins in Arabidopsis. J. Exp. Bot. 2016, 67, 5203–5215. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chalkley, R.; Maynard, J.; Wang, W.; Ni, W.; Jiang, X.; Shin, K.; Cheng, L.; Savage, D.; Hühmer, A.; et al. Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Benschop, J.; Mohammed, S.; O’Flaherty, M.; Heck, A.; Slijper, M.; Menke, F. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteom. 2007, 6, 1198–1214. [Google Scholar] [CrossRef] [PubMed]
- Nespoulous, C.; Rofidal, V.; Sommerer, N.; Hem, S.; Rossignol, M. Phosphoproteomic analysis reveals major default phosphorylation sited outside mong intrinsically disordered regions of Arabidopsis plasma membrane proteins. Proteome Sci. 2012, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Nühse, T.; Stensballe, A.; Jensen, O.; Peck, S. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 2004, 16, 2394–2405. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Hwang, B. The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 2014, 155, 447–463. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Aslanian, A.; Yates, J., III. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008, 12, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, J.B.; Nasrallah, M. S-locus receptor kinase signalling. Biochem. Soc. Trans. 2014, 42, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Deb, S.; Sankaranarayanan, S.; Wewala, G.; Widdup, E.; Samuel, M. The S-domain receptor kinase Arabidopsis receptor kinase2 and the U box/armadillo repeat-containing E3 ubiquitin ligase9 module mediates lateral root development under phosphate starvation in Arabidopsis. Plant Physiol. 2014, 165, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Trontin, C.; Kiani, S.; Corwin, J.; Hematy, K.; Yansouni, J.; Kliebenstein, D.; Loudet, O. A pair of receptor-like kinases is responsible for natural variation in shoot growth response to mannitol treatment in Arabidopsis thaliana. Plant J. 2014, 78, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S.; Gisch, N.; Schaffer, M.; Illig, T.; Westphal, L.; Knirel, Y.; Sanchez-Carballo, P.; Zahringer, U.; Huckelhoven, R.; Lee, J.; et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 2015, 16, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G.; et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jung, M.; Lee, S.; Kim, K.; Byun, H.; Choi, M.; Park, H.; Cho, M.; Chung, W. An S-locus receptor-like kinase plays a role as a negative regulator in plant defense responses. Biochem. Biophys. Res. Commun. 2009, 381, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Gilardoni, P.; Hettenhausen, C.; Baldwin, I.; Bonaventure, G. Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell Environ. 2011, 23, 3512–3532. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wu, Y.; Guo, J.; Du, B.; Chen, R.; Zhu, L.; He, G. A rice lectin receptor-like kinase is involved in innate immune responses also contributes to seed germination. Plant J. 2013, 76, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.; Diener, A. Diversity in receptor-like kinase genes is a major determinant of quantitative resistance to Fusarium oxysporum f.sp. matthioli. New Phytol. 2013, 200, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, H.; Chen, H.; Liu, Y.; He, J.; Kang, H.; Sun, Z.; Pan, G.; Wang, Q.; Hu, J.; et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 2015, 33, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Guidarelli, M.; Zoli, L.; Orlandini, A.; Bertolini, P.; Baraldi, E. The mannose-binding lectin gene FaMBL1 is involved in the resistance of unripe strawberry fruits to Colletotrichum acutatum. Mol. Plant Pathol. 2014, 15, 832–840. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, R.; Fobis-Loisy, I.; Gaude, T. When no means no: Guide to Brassicaceae self-incompatibility. Trends Plant Sci. 2010, 15, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, M.; Liu, P.; Sherman-Broyles, S.; Boggs, N.; Nasrallah, J. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: Implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 2004, 101, 16070–16074. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, M.; Liu, P.; Sherman-Broyles, S.; Boggs, N.; Nasrallah, J. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 2002, 297, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Boggs, N.; Dwyer, K.; Nasrallah, M.; Nasrallah, J. In vivo detection of residues required for ligand-selective activation of the S-locus receptor kinase in Arabidopsis. Curr. Biol. 2009, 19, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Tantikanjana, T.; Nishio, T.; Nasrallah, M.; Nasrallah, J. Site-specific N-glycosylation of the S-locus receptor kinase and its role in the self-incompatibility response of the brassicaceae. Plant Cell 2014, 26, 4749–4762. [Google Scholar] [CrossRef] [PubMed]
- Naithani, S.; Chookajorn, T.; Ripoll, D.; Nasrallah, J. Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl. Acad. Sci. USA 2007, 104, 12211–12216. [Google Scholar] [CrossRef] [PubMed]
- Shimosato, H.; Yokota, N.; Shiba, H.; Iwano, M.; Entani, T.; Che, F.; Watanabe, M.; Isogai, A.; Takayama, S. Characterization of the SP11/SCR high affinity binding site involved in self/non self recognition in brassica self-incompatibility. Plant Cell 2007, 19, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Kakita, M.; Murase, K.; Iwano, M.; Matsumoto, T.; Watanabe, M.; Shiba, H.; Isogai, H.; Takayama, S. Two distinct forms of M-locus protein kinase localize at the plasma membrane and interact directly with S-locus receptor kinase to transducer self-incompatibility signalling in Brassica rapa. Plant Cell 2007, 19, 3961–3973. [Google Scholar] [CrossRef] [PubMed]
- Indriolo, E.; Tharmapalan, P.; Wright, S.; Goring, D. The ARC1 E3 ligase gene is frequently deleted in self-compatible Brassicaceae species ans has a conserved role in Arabidopsis lyrata self-pollen rejection. Plant Cell 2012, 24, 4607–4620. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.; Anderson, E.; Mullen, R.; Goring, D. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 2003, 15, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.; Mudgil, Y.; Salt, J.; Delmas, F.; Ramachandran, S.; Chilelli, A.; Goring, D. Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 2008, 147, 2084–2095. [Google Scholar] [CrossRef] [PubMed]
- Gouget, A.; Senchou, V.; Govers, F.; Sanson, A.; Barre, A.; Rouge, P.; Pont-Lezica, R.; Canut, H. Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol. 2006, 140, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Govers, F. Arabidopsis L-type lectin receptor kinases: Phylogeny, classification, and expression profiles. J. Exp. Bot. 2009, 60, 4383–4396. [Google Scholar] [CrossRef] [PubMed]
- Gouhier-Darimont, C.; Schmiesing, A.; Bonnet, C.; Lassueur, S.; Reymond, P. Signalling of Arabidopsis thaliana response to Pieris brassicae eggs shares similarities with PAMP-triggered immunity. J. Exp. Bot. 2013, 64, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Balagué, C.; Gouget, A.; Bouchez, O.; Souriac, C.; Haget, N.; Boutet-Mercey, S.; Govers, F.; Roby, D.; Canut, H. The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Yeh, Y.; Liu, A.; Cheng, C.; Zimmerli, L. The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 2014, 79, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kuo, Y.C.; Mishra, S.; Tsai, C.; Chien, C.; Chen, C.; Desclos-Theveniau, M.; Chu, P.; Schulze, B.; Chinchilla, D.; et al. The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 2012, 24, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Desclos-Theveniau, M.; Arnaud, D.; Huang, T.; Lin, G.; Chen, W.; Lin, Y.; Zimmerli, L. The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 2012, 8, e1002513. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cordewener, J.; America, A.; Shan, W.; Bouwmeester, K.; Govers, F. Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 are functional analogs in regulating phytophthora resistance and plant cell death. Mol. Plant Microbe Interact. 2015, 28, 1032–1048. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; de Sain, M.; Weide, R.; Gouget, A.; Klamer, S.; Canut, H.; Govers, F. The lectin kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011, 7, e1001327. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nsibo, D.; Juhar, H.M.; Govers, F.; Bouwmeester, K. Ectopic expression of Arabidopsis L-type lectin receptor kinase genes LecRK-I.9 and LecRK-IX.1 in Nicotiana benthamiana confers Phytophthora resistance. Plant Cell Rep. 2016, 35, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Ju, H.; Min, J.; Zhang, X.; Kim, S.; Yang, K.; Kim, C. Overexpression of L-type lectin-like protein kinase 1 confers pathogen resistance and regulates salinity response in Arabidopsis thaliana. Plant Sci. 2013, 203–204, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bouwmeester, K.; Beseh, P.; Shan, W.; Govers, F. Phenotypic analyses of Arabidopsis T-DNA insertion lines and expression profiling reveal that multiple L-type lectin receptor kinases are involved in plant immunity. Mol. Plant Microbe Interact. 2014, 27, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Patel, A.; Mathieu, M.; Kim, S.; Xu, D.; Stacey, G. A lectin receptor-like kinase is required for pollen development in Arabidopsis. Plant Mol. Biol. 2008, 67, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Wang, A.; Yang, G.; Gao, P.; Zheng, Z. The Arabidopsis A4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination. Plant Physiol. 2009, 149, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Canut, H.; Carrasco, A.; Galaud, J.; Cassan, C.; Bouyssou, H.; Vita, N.; Ferrara, P.; Pont-Lezica, R. High affinity RGD-binding sites at the plasma membrane of Arabidopsis thaliana links the cell wall. Plant J. 1998, 16, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Mellersh, D.; Heath, M. Plasma-membrane cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 2001, 13, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Senchou, V.; Weide, R.; Carrasco, A.; Bouyssou, H.; Pont-Lezica, R.; Govers, F.; Canut, H. High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell. Mol. Life Sci. 2004, 61, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Choi, J.; Cao, Y.; Stacey, G. Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front. Plant Sci. 2014, 5, 446. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Yoshida, R.; Ichimura, K.; Mizoguchi, T.; Seo, S.; Yonezawa, M.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 2007, 19, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.; Tanaka, K.; Cao, Y.; Cho, S.; Xu, D.; Stacey, G. Computational analysis of the ligand binding site of the extracellular ATP receptor, DORN1. PLoS ONE 2016, 11, e0161894. [Google Scholar] [CrossRef] [PubMed]
- André, S.; Siebert, H.; Nishiguchi, M.; Tazaki, K.; Gabius, H. Evidence for lectin activity of a plant receptor-like protein kinase by application of neoglycoproteins and bioinformatic algorithms. Biochim. Biophys. Acta 2005, 1725, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Adar, R.; Sharon, N. Mutational studies of the amino acid residues in the combining site of Erythrina corallodendron lectin. Eur. J. Biochem. 1996, 239, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chakraborty, S.; Xu, G. X-ray crystallographic studies of the extracellular domain of the first plant ATP receptor, DORN1, and the orthologous protein from Camelina sativa. Acta Cryst. 2016, F72, 782–787. [Google Scholar]
- Singh, P.; Zimmerli, L.; Singh, P.; Zimmerli, L. Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 2014, 4, 124. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Van Damme, E. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014, 5, 397. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ramonell, K.; Somerville, S.; Stacey, G. Characterization of early, chitin-induced gene expression in Arabidopsis. Mol. Plant Microbe Interact. 2002, 15, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Lyou, S.; Park, H.; Jung, C.; Sohn, H.; Lee, G.; Kim, C.; Kim, M.; Choi, Y.; Cheong, J. The Arabidopsis AtLEC gene encoding a lectin-like protein is up-regulated by multiple stimuli including developmental signal, wounding, jasmonate, ethylene, and chitin elicitor. Mol. Cells 2009, 27, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Armijo, G.; Salinas, P.; Monteoliva, M.; Seguel, A.; Garcia, C.; Villarroel-Candia, E.; Song, W.; van der Krol, A.; Alvarez, M.; Holuigue, L. A salicylic acid-induced lectin-like protein plays a positive role in the effector-triggered immunity response of Arabidopsis thaliana to Pseudomonas syringae Avr-Rpm1. Mol. Plant Microbe Interact. 2013, 26, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Brill, L.; Evans, C.; Hirsch, A. Expression of MsLEC1- and MsLEC2-antisense genes in alfalfa plant lines causes severe embryogenic, developmental and reproductive abnormalities. Plant J. 2001, 25, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Joris, B.; Englebert, S.; Chu, C.-P.; Kariyama, R.; Daneo-Moore, L.; Shockman, G.; Ghuysen, J.-M. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 1992, 91, 257–264. [Google Scholar] [CrossRef]
- Ponting, C.; Aravind, L.; Schultz, J.; Bork, P.; Koonin, E. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 1999, 289, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Bycroft, M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 2000, 299, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.; Madsen, E.; Felle, H.; Umehara, Y.; Gronlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.; Madsen, L.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.; Willmann, R.; Desaki, Y.; Grabherr, H.; Nürnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Oldroyd, G. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Kawaharada, Y.; Kelly, S.; Nielsen, M.; Hjuler, C.; Gysel, K.; Muszynski, A.; Carlson, R.; Thygesen, M.; Sandal, N.; Asmussen, M.; et al. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 2015, 523, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [PubMed]
- Iizasa, E.; Mitsutomi, M.; Nagano, Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J. Biol. Chem. 2010, 285, 2996–3004. [Google Scholar] [CrossRef] [PubMed]
- Petutschnig, E.; Jones, A.; Serazetdinova, L.; Lipka, U.; Lipka, V. The Lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 2010, 285, 28902–28911. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Tanaka, K.; Zhang, X.-C.; Son, G.; Brechenmacher, L.; Nguyen, T.; Stacey, G. LYK4, a Lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012, 160, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.; Jedrzejczak, R.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, C.; Petutschnig, E.; Benitez-Alfonso, Y.; Beck, M.; Robatzek, S.; Lipka, V.; Maule, A. LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. USA 2013, 110, 9166–9170. [Google Scholar] [CrossRef] [PubMed]
- Willmann, R.; Lajunen, H.; Erbs, G.; Newman, M.-A.; Kolb, D.; Tsuda, K.; Katagiri, F.; Fliegmann, J.; Bono, J.-J.; Cullimore, J.; et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2011, 108, 19824–19829. [Google Scholar] [CrossRef] [PubMed]
- Hayafune, M.; Berisio, R.; Marchetti, R.; Silipo, A.; Kayama, M.; Desaki, Y.; Arima, S.; Squeglia, F.; Ruggiero, A.; Tokuyasu, K.; et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. USA 2014, 111, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Kouzai, Y.; Nakajima, K.; Hayafune, M.; Ozawa, K.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol. Biol. 2014, 84, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, J.; Han, Z.; Gong, X.; Zhang, H.; Chai, J. Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure 2016, 24, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, J.-F.; Ao, Y.; Qu, J.; Li, Z.; Su, J.; Zhang, Y.; Liu, J.; Feng, D.; Qi, K.; et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 2012, 24, 3406–3419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, W.; Sun, J.; Feng, F.; Deng, Y.; He, Z.; Oldroyd, G.; Wang, E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015, 81, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; van Zeijl, A.; Geurts, R. Lipochitooligosaccharides modulate plant host immunity to enable endosymbioses. Annu. Rev. Phytopathol. 2015, 53, 311–334. [Google Scholar] [CrossRef] [PubMed]
- Pietraszewska-Bogiel, A.; Lefebvre, B.; Koini, M.; Klaus-Heisen, D.; Takken, F.; Cullimore, J.; Gadella, T. Interaction of Medicago truncatula Lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana leaf induces defence-like responses. PLoS ONE 2013, 8, e65055. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.; Antolín-Llovera, M.; Grossmann, C.; Ye, J.; Vieweg, S.; Broghammer, A.; Krusell, L.; Radutoiu, S.; Jensen, O.; Stougaard, J.; et al. Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 2011, 65, 404–417. [Google Scholar] [CrossRef] [PubMed]
- Gough, C.; Jacquet, C. Nod factor perception protein carries weight in biotic interactions. Trends Plant Sci. 2013, 18, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Gourion, B.; Berrabah, F.; Ratet, P.; Stacey, G. Rhizobium-legume symbioses: The crucial role of plant immunity. Trends Plant Sci. 2015, 20, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Cao, Y.; Tanaka, K.; Thibivilliers, S.; Wan, J.; Choi, J.; Kang, C.; Qiu, J.; Stacey, G. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 2013, 341, 1384–1387. [Google Scholar] [CrossRef] [PubMed]
- Stracke, T.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Endre, G.; Kereszt, A.; Kevei, Z.; Mihacea, S.; Kalo, P.; Kiss, G. A receptor kinase gene regulating symbiotic nodule development. Nature 2002, 417, 962–966. [Google Scholar] [CrossRef] [PubMed]
- Antolín-Llovera, M.; Ried, M.; Parniske, M. Cleavage of the symbiosis receptor-like kinase ectodomain promotes complex formation with Nod factor receptor 5. Curr. Biol. 2014, 24, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Schallus, T.; Jaeckh, C.; Fehér, K.; Palma, A.; Liu, Y.; Simpson, J.; Mackeen, M.; Stier, G.; Gibson, T.; Feizi, T.; et al. Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 2008, 19, 3404–3414. [Google Scholar] [CrossRef] [PubMed]
- Schallus, T.; Fehér, K.; Sternberg, U.; Rybin, V.; Muhle-Goll, C. Analysis of the specific interactions between the lectin domain of malectin and diglucosides. Glycobiology 2010, 20, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Hok, S.; Allasia, V.; Andrio, E.; Naessens, E.; Ribes, E.; Panabières, F.; Attard, A.; Ris, N.; Clement, M.; Barlet, X.; et al. The receptor kinase IMPAIRED OOMYCETE SUSCEPTIBILITY1 attenuates abscisic acid responses in Arabidopsis. Plant Physiol. 2014, 166, 1506–1518. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.; Panzeri, D.; Kadota, Y.; Huang, Y.; Huang, P.; Tao, C.; Roux, M.; Chien, H.; Chin, T.; Chu, P.; et al. The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 2016, 28, 1701–1721. [Google Scholar] [CrossRef] [PubMed]
- Pagnussat, G.; Yu, H.-J.; Ngo, Q.; Rajani, S.; Mayalagu, S.; Johnson, C.; Capron, A.; Xie, L.-F.; Ye, D.; Sundaresan, V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 2005, 132, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.; Müller, L.; Boisson-Dernier, A.; Grossniklaus, U.T. CrRLK1L receptor-like kinases: Not just another brick in the wall. Curr. Opin. Plant Biol. 2012, 15, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Hematy, K.; Sado, P.-E.; Van Tuinen, A.; Rochange, S.; Desnos, T.; Balzergue, S.; Pelletier, S.; Renou, J.-P.; Höfte, H. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 2007, 17, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Li, L.; Ye, H.; Yu, X.; Algreen, A.; Yin, Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 7648–7653. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ye, H.; Li, L.; Yin, Y. A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signal. Behav. 2009, 4, 784–786. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.; Wu, H. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dernier, A.; Roy, S.; Kritsas, K.; Grobei, M.; Jaciubek, M.; Schroeder, J.; Grossniklaus, U. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 2009, 136, 3279–3288. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, S.; Murata, T.; Sakurai-Ozato, N.; Kubo, M.; Demura, T.; Fukuda, H.; Hasebe, M. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 2009, 19, 1327–1331. [Google Scholar] [CrossRef] [PubMed]
- Huck, N.; Moore, J.; Federer, M.; Grossniklaus, U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 2003, 130, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Rotman, N.; Rozier, F.; Boavida, L.; Dumas, C.; Berger, F.; Faure, J. Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 2003, 13, 432–436. [Google Scholar] [CrossRef]
- Kanaoka, M.; Torii, K. FERONIA as an upstream receptor kinase for polar growth in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 17461–17462. [Google Scholar] [CrossRef] [PubMed]
- Knight, M. New ideas on root hair growth appear from the flanks. Proc. Natl. Acad. Sci. USA 2007, 104, 10649–10650. [Google Scholar] [CrossRef] [PubMed]
- Francoz, E.; Ranocha, P.; Nguyen-Kim, H.; Jamet, E.; Burlat, V.; Dunand, C. Roles of cell wall peroxidases in plant development. Phytochemistry 2015, 112, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Kita, D.; Johnson, E.; Aggarwal, M.; Gates, L.; Wu, H.; Cheung, A. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 2014, 5, 3129. [Google Scholar] [CrossRef] [PubMed]
- Denness, L.; McKenna, J.; Segonzac, C.; Wormit, A.; Madhou, P.; Bennett, M.; Mansfield, J.; Zipfel, C.; Hamann, T. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen spacies- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 2011, 156, 1364–1374. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dernier, A.; Lituiev, D.; Nestorova, A.; Franck, C.; Thirugnanarajah, S.; Grossniklaus, U. ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol. 2013, 11, e1001719. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhou, Y.; Ma, X.; Gao, L.; Song, C. Arabidopsis CAP1-mediated ammonium sensing required reactive oxygen species in plant cell growth. Plant Signal. Behav. 2014, 9, e29582. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.; Sussman, M. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Li, X.; Chen, J.; Chen, W.; Li, B.; Li, C.; Wang, L.; Li, J.; Zhao, X.; Lin, J.; et al. Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 8326–8334. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.; Miller, N.; Dai, C.; Spalding, E.; Monshausen, G. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr. Biol. 2014, 24, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Qian, L.; Nibau, C.; Duan, Q.; Kita, D.; Levasseur, K.; Li, X.; Lu, C.; Li, H.; Hou, C.; et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 14693–14698. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, F.; Liu, Y.; Du, C.; Li, X.; Zhu, S.; Wang, X.; Lan, W.; Rodriguez, P.; Liu, X.; et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 5519–5527. [Google Scholar] [CrossRef] [PubMed]
- Nühse, T. Cell wall integrity and innate immunity in plants. Front. Plant Sci. 2012, 3, 280. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signalling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Kessler, S.; Shimosato-Asano, H.; Keinath, N.; Wuest, S.; Ingram, G.; Panstruga, R.; Grossniklaus, U. Conserved molecular components for pollen tube reception and fungal invasion. Science 2010, 330, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Govers, F.; Angenent, G. Fertility goddesses as Trojan horses. Science 2010, 330, 922–923. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.; Kessler, S.; Müller, L.; Shimosato-Asano, H.; Boisson-Dernier, A.; Grossniklaus, U. TURAN and EVAN mediate pollen tube reception in Arabidopsis synergids through protein glycosylation. PLoS Biol. 2015, 13, e1002139. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Höfte, H. Growth control: A saga of cell walls, ROS, and peptide receptors. Plant Cell Environ. 2014, 26, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.; Fangel, J.; McCleary, B.; Ruzanski, C.; Rydahl, M.; Ralet, M.; Farkas, V.; von Schantz, L.; Marcus, S.; Andersen, M.; et al. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J. Biol. Chem. 2012, 287, 39429–39438. [Google Scholar] [CrossRef] [PubMed]
- Mallèvre, F.; Roget, A.; Kervella, Y.; Ropartz, D.; Ralet, M.; Canut, H.; Minon, T.; Livache, T. Microwave heating for the rapid generation of glycosylhydrazides. Bioconjug. Chem. 2013, 24, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Lease, K.A.; Walker, J.C. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 2006, 142, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Smith, S.; De Smet, I. Small signaling peptides in Arabidopsis development: How cells communicate over a short distance. Plant Cell 2012, 24, 3198–3217. [Google Scholar] [CrossRef] [PubMed]
- Nabi, I.; Shankar, J.; Dennis, J. The galectins lattice at a glance. J. Cell Sci. 2015, 128, 2213–2219. [Google Scholar] [CrossRef] [PubMed]
- Elola, M.; Blidner, A.; Ferragut, F.; Bracalente, C.; Rabinovich, G. Assembly, organization and regulation of cell-surface receptors by lectin-glycan complexes. Biochem. J. 2015, 469, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Irani, N.; Russinova, E. Receptor endocytosis and signaling in plants. Curr. Opin. Plant Biol. 2009, 12, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rubio, M.; Alassimone, J.; Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 2013, 153, 402–412. [Google Scholar] [CrossRef] [PubMed]





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellande, K.; Bono, J.-J.; Savelli, B.; Jamet, E.; Canut, H. Plant Lectins and Lectin Receptor-Like Kinases: How Do They Sense the Outside? Int. J. Mol. Sci. 2017, 18, 1164. https://doi.org/10.3390/ijms18061164
Bellande K, Bono J-J, Savelli B, Jamet E, Canut H. Plant Lectins and Lectin Receptor-Like Kinases: How Do They Sense the Outside? International Journal of Molecular Sciences. 2017; 18(6):1164. https://doi.org/10.3390/ijms18061164
Chicago/Turabian StyleBellande, Kevin, Jean-Jacques Bono, Bruno Savelli, Elisabeth Jamet, and Hervé Canut. 2017. "Plant Lectins and Lectin Receptor-Like Kinases: How Do They Sense the Outside?" International Journal of Molecular Sciences 18, no. 6: 1164. https://doi.org/10.3390/ijms18061164