Circulating miRNA-192 and miR-29a as Disease Progression Biomarkers in Hepatitis C Patients with a Prevalence of HCV Genotype 3
Abstract
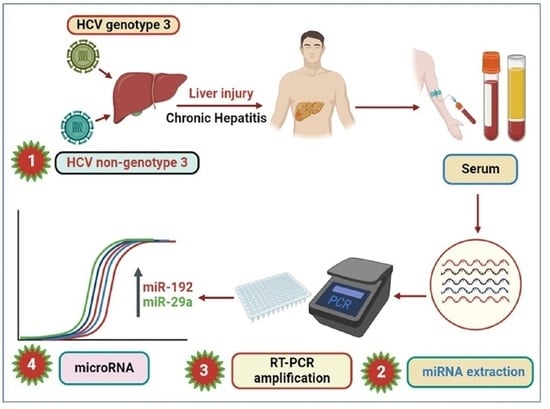
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Isolation
2.3. Synthesis of cDNA and RT-PCR
2.4. Data Analysis
3. Results
3.1. Characteristics of the Patient Cohort
3.2. Serum miR-192 and miR-29a Profile
3.3. Correlation of miR-192 and 29a with ALT
3.4. Expression of miR-192 and miR-29a in HCV Genotypes 3 and Non-Genotypes 3
3.5. ROC Analysis in miR-192 and miR-29a in the HCV Genotypes 3 and Non-Genotypes 3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- AbdElrazek, S.E.; Bilasy, A.E.; AbdElhalim, A.E. Prior to the oral therapy, what do we know about HCV-4 in Egypt: A randomized survey of prevalence and risks using data mining computed analysis. Medicine 2014, 93, e204. [Google Scholar] [CrossRef]
- Lanini, S.; Ustianowski, A.; Pisapia, R.; Zumla, A.; Ippolito, G. Viral hepatitis: Etiology, epidemiology, transmission, diagnostics, treatment, and prevention. Infect. Dis. Clin. 2019, 33, 1045–1062. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Rehman, I.U.; Ahmad, J.; Gohar, M.; Ahmad, S.; Ahmad, B. Hepatitis-C Virus and Cirrhosis: An Overview from Khyber Pakhtunkhwa Province of Pakistan. Viral Immunol. 2020, 33, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Idrees, M.; Riazuddin, S. Frequency distribution of hepatitis C virus genotypes in different geographical regions of Pakistan and their possible routes of transmission. BMC Infect. Dis. 2008, 8, 69. [Google Scholar] [CrossRef]
- Butt, S.; Idrees, M.; Akbar, H.; Rehman, I.U.; Awan, Z.; Afzal, S.; Hussain, A.; Shahid, M.; Manzoor, S.; Rafique, S. The changing epidemiology pattern and frequency distribution of hepatitis C virus in Pakistan. Infect. Genet. Evol. 2010, 10, 595–600. [Google Scholar] [CrossRef]
- Gonzalez, H.C.; Duarte-Rojo, A. Virologic cure of hepatitis C: Impact on hepatic fibrosis and patient outcomes. Curr. Gastroenterol. Rep. 2016, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, L.E.; Gambardella, M.; Andreana, A.; Tripodi, M.F.; Utili, R.; Ruggiero, G. Steatosis accelerates the progression of liver damage of chronic hepatitisC patients and correlates with specific HCV genotype and visceral obesity. Hepatology 2001, 33, 1358. [Google Scholar] [CrossRef]
- Loosen, S.H.; Schueller, F.; Trautwein, C.; Roy, S.; Roderburg, C. Role of circulating microRNAs in liver diseases. World J. Hepatol. 2017, 9, 586. [Google Scholar] [CrossRef]
- Schueller, F.; Roy, S.; Vucur, M.; Trautwein, C.; Luedde, T.; Roderburg, C. The Role of miRNAs in the Pathophysiology of Liver Diseases and Toxicity. Int. J. Mol. Sci. 2018, 19, 261. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From mechanism to organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- Liang, H.; Gong, F.; Zhang, S.; Zhang, C.Y.; Zen, K.; Chen, X. The origin, function, and diagnostic potential of extracellular microRNAs in human body fluids. Wiley Interdiscip. Rev. RNA 2014, 5, 285–300. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Bueno, M.J.; de Castro, I.P.; Malumbres, M. Control of cell proliferation pathways by microRNAs. Cell Cycle 2008, 7, 3143–3148. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, J.H.; Lee, S.W. The role of microRNAs in hepatitis C virus replication and related liver diseases. J. Microbiol. 2014, 52, 445–451. [Google Scholar] [CrossRef]
- Yu, X.; Odenthal, M.; Fries, J.W. Exosomes as miRNA Carriers: Formation-Function-Future. Int. J. Mol. Sci. 2016, 17, 2028. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Manriquez, L.M.; Carrasco-Morales, O.; Osorio-Perez, S.M.; Estrada-Meza, C.; Pathak, S.; Banerjee, A.; Bandyopadhyay, A.; Duttaroy, A.K.; Paul, S. MicroRNA-mediated regulation of key signaling pathways in hepatocellular carcinoma: A mechanistic insight. Front. Genet. 2022, 13, 910733. [Google Scholar] [CrossRef] [PubMed]
- Morishita, A.; Oura, K.; Tadokoro, T.; Fujita, K.; Tani, J.; Masaki, T. MicroRNAs in the pathogenesis of hepatocellular carcinoma: A review. Cancers 2021, 13, 514. [Google Scholar] [CrossRef]
- Cabral, B.; Hoffmann, L.; Bottaro, T.; Costa, P.; Ramos, A.; Coelho, H.; Villela-Nogueira, C.; Ürményi, T.; Faffe, D.; Silva, R. Circulating microRNAs associated with liver fibrosis in chronic hepatitis C patients. Biochem. Biophys. Rep. 2020, 24, 100814. [Google Scholar] [CrossRef]
- Trebicka, J.; Anadol, E.; Elfimova, N.; Strack, I.; Roggendorf, M.; Viazov, S.; Wedemeyer, I.; Drebber, U.; Rockstroh, J.; Sauerbruch, T.; et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J. Hepatol. 2013, 58, 234. [Google Scholar] [CrossRef]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef]
- Kumar, A.; Rajput, M.K.; Paliwal, D.; Yadav, A.; Chhabra, R.; Singh, S. Genotyping & diagnostic methods for hepatitis C virus: A need of low-resource countries. Indian J. Med. Res. 2018, 147, 445. [Google Scholar]
- Cabrera, M.; Kolli, M.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. miRNA-205: A future therapeutic molecule for liver diseases. Future Drug Discov. 2023, 4, FDD78. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Elfimova, N.; Müller, M.; Bachurski, D.; Koitzsch, U.; Drebber, U.; Mahabir, E.; Hansen, H.P.; Friedman, S.L.; Klein, S.; et al. Autophagy-Related Activation of Hepatic Stellate Cells Reduces Cellular miR-29a by Promoting Its Vesicular Secretion. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 1701–1716. [Google Scholar] [CrossRef]
- Noetel, A.; Kwiecinski, M.; Elfimova, N.; Huang, J.; Odenthal, M. microRNA are central players in anti-and profibrotic gene regulation during liver fibrosis. Front. Physiol. 2012, 3, 49. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [PubMed]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Luedde, T. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, C.H.; Lee, S.W. Hepatitis C virus infection stimulates transforming growth factor-β1 expression through up-regulating miR-192. J. Microbiol. 2016, 54, 520–526. [Google Scholar] [CrossRef]
- Liu, X.-L.; Pan, Q.; Cao, H.-X.; Xin, F.-Z.; Zhao, Z.-H.; Yang, R.-X.; Zeng, J.; Zhou, H.; Fan, J.-G. Lipotoxic hepatocyte-derived exosomal microRNA 192-5p activates macrophages through rictor/Akt/forkhead box transcription factor O1 signaling in nonalcoholic fatty liver disease. Hepatology 2020, 72, 454–469. [Google Scholar] [CrossRef]
- Ezaz, G.; Trivedi, H.D.; Connelly, M.A.; Filozof, C.; Howard, K.; Parrish, M.; Kim, M.; Herman, M.A.; Nasser, I.; Afdhal, N.H.; et al. Differential associations of circulating MicroRNAs with pathogenic factors in NAFLD. Hepatol. Commun. 2020, 4, 670–680. [Google Scholar] [CrossRef]
- Ullah, A.; Yu, X.; Odenthal, M.; Meemboor, S.; Ahmad, B.; Rehman, I.U.; Ahmad, J.; Ali, Q.; Nadeem, T. Circulating microRNA-122 in HCV cirrhotic patients with high frequency of genotype 3. PLoS ONE 2022, 17, e0268526. [Google Scholar] [CrossRef]
- Anadol, E.; Schierwagen, R.; Elfimova, N.; Tack, K.; Schwarze-Zander, C.; Eischeid, H.; Noetel, A.; Boesecke, C.; Jansen, C.; Dold, L.; et al. Circulating microRNAs as a marker for liver injury in human immunodeficiency virus patients. Hepatology 2015, 61, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.-J.; Yao, Y.; Cai, X.-Y.; Fang, G.-Y. Emerging Role of MiR-192-5p in Human Diseases. Front. Pharmacol. 2021, 12, 614068. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Benz, F.; Alder, J.; Bantel, H.; Janssen, J.; Vucur, M.; Gautheron, J.; Schneider, A.; Schüller, F.; Loosen, S.; et al. Downregulation of miR-192-5p protects from oxidative stress-induced acute liver injury. Clin. Sci. 2016, 130, 1197–1207. [Google Scholar] [CrossRef]
- Motawi, T.K.; Shaker, O.G.; El-Maraghy, S.A.; Senousy, M.A. Serum interferon-related microRNAs as biomarkers to predict the response to interferon therapy in chronic hepatitis C genotype 4. PLoS ONE 2015, 10, e0120794. [Google Scholar] [CrossRef]
- Mahdy, M.M.; El-Ekiaby, N.M.; Hashish, R.M.; Salah, R.A.; Hanafi, R.; Azzazy, H.M.E.-S.; Abdelaziz, A.I. miR-29a promotes lipid droplet and triglyceride formation in HCV infection by inducing expression of SREBP-1c and CaV1. J. Clin. Transl. Hepatol. 2016, 4, 293. [Google Scholar] [PubMed]
- Bandyopadhyay, S.; Friedman, R.C.; Marquez, R.T.; Keck, K.; Kong, B.; Icardi, M.S.; Brown, K.E.; Burge, C.B.; Schmidt, W.N.; Wang, Y.; et al. Hepatitis c virus infection and hepatic stellate cell activation downregulate mir-29: Mir-29 overexpression reduces hepatitis c viral abundance in culture. J. Infect. Dis. 2011, 203, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Lee, Y.; Lee, Y.-S.; Gim, J.-A.; Ko, E.; Yim, S.Y.; Jung, Y.K.; Kang, S.; Kim, M.Y.; Kim, H.; et al. Circulating mirna is a useful diagnostic biomarker for nonalcoholic steatohepatitis in nonalcoholic fatty liver disease. Sci. Rep. 2021, 11, 14639. [Google Scholar] [CrossRef]
- Iacob, D.G.; Rosca, A.; Ruta, S. Circulating micrornas as non-invasive biomarkers for hepatitis b virus liver fibrosis. World J. Gastroenterol. 2020, 26, 1113–1127. [Google Scholar] [CrossRef]






| S. No. | Area under the ROC Curve | 95% Confidence Interval | Std. Error | p Value |
|---|---|---|---|---|
| Control–Mild genotype 3 | 0.6549 | 0.4763 to 0.8335 | 0.09108 | 0.2564 |
| Control–Mild non-genotype 3 | 0.6356 | 0.3783 to 0.8929 | 0.1312 | 0.2806 |
| Control–Moderate genotype 3 | 0.7276 | 0.5866 to 0.8686 | 0.07193 | <0.001829 |
| Control–Moderate non-genotype 3 | 0.8687 | 0.7674 to 0.9700 | 0.05168 | <0.0001 |
| Control–Severe genotype 3 | 0.5176 | 0.3789 to 0.6562 | 0.07073 | 0.8071 |
| Control–Severe non-genotype 3 | 0.5438 | 0.3880 to 0.6996 | 0.07949 | 0.6083 |
| Control vs. Genotype 3 | 0.6237 | 0.5152 to 0.7321 | 0.05532 | <0.03055 |
| Control vs. Non-genotype 3 | 0.7079 | 0.5987 to 0.8171 | 0.05569 | <0.0007660 |
| Genotype 3 vs. Non-genotype 3 | 0.5816 | 0.4625 to 0.7007 | 0.06076 | 0.1845 |
| S. No. | Area under the ROC Curve | 95% Confidence Interval | Std. Error | p Value |
|---|---|---|---|---|
| Control–Mild genotype 3 | 0.7200 | 0.4736 to 0.9663 | 0.1256 | <0.04667 |
| Control–Mild non-genotype 3 | 0.7981 | 0.6735 to 0.9227 | 0.06357 | <0.002033 |
| Control–Moderate genotype 3 | 0.7800 | 0.6703 to 0.8898 | 0.05596 | <0.0001 |
| Control–Moderate non-genotype 3 | 0.5971 | 0.4759 to 0.7184 | 0.06186 | 0.1142 |
| Control–Severe genotype 3 | 0.7618 | 0.6461 to 0.8776 | 0.05905 | <0.0001772 |
| Control–Severe non-genotype 3 | 0.6945 | 0.5969 to 0.7920 | 0.04976 | 0.0002866 |
| Control vs. Genotype 3 | 0.7636 | 0.6728 to 0.8544 | 0.04631 | <0.0001 |
| Control vs. Non-genotype 3 | 0.6718 | 0.5852 to 0.7583 | 0.04416 | 0.0003750 |
| Genotype 3 vs. Non-genotype 3 | 0.6102 | 0.5206 to 0.6997 | 0.04568 | 0.01785 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, A.; Rehman, I.U.; Ommer, K.; Ahmed, N.; Odenthal, M.; Yu, X.; Ahmad, J.; Nadeem, T.; Ali, Q.; Ahmad, B. Circulating miRNA-192 and miR-29a as Disease Progression Biomarkers in Hepatitis C Patients with a Prevalence of HCV Genotype 3. Genes 2023, 14, 1056. https://doi.org/10.3390/genes14051056
Ullah A, Rehman IU, Ommer K, Ahmed N, Odenthal M, Yu X, Ahmad J, Nadeem T, Ali Q, Ahmad B. Circulating miRNA-192 and miR-29a as Disease Progression Biomarkers in Hepatitis C Patients with a Prevalence of HCV Genotype 3. Genes. 2023; 14(5):1056. https://doi.org/10.3390/genes14051056
Chicago/Turabian StyleUllah, Amin, Irshad Ur Rehman, Katharina Ommer, Nadeem Ahmed, Margarete Odenthal, Xiaojie Yu, Jamshaid Ahmad, Tariq Nadeem, Qurban Ali, and Bashir Ahmad. 2023. "Circulating miRNA-192 and miR-29a as Disease Progression Biomarkers in Hepatitis C Patients with a Prevalence of HCV Genotype 3" Genes 14, no. 5: 1056. https://doi.org/10.3390/genes14051056