Superfast Gelation of Spider Silk-Based Artificial Silk Protein
Abstract
:1. Introduction
2. Results and Discussion
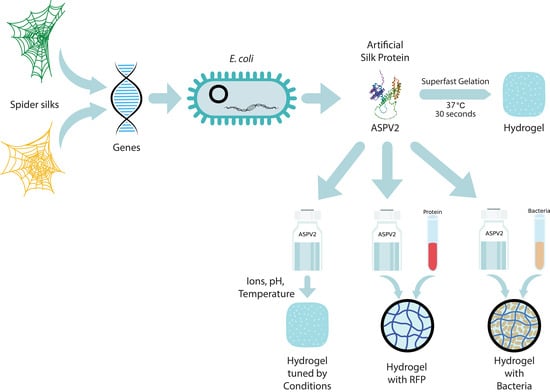
2.1. Fabrication of Hydrogels from Genetically Engineered Artificial Silk Proteins
2.2. Superfast Gelation Efficiency of Engineered Artificial Silk Protein ASPV2
2.3. Modulation of ASPV2 Gelation Time by Ionic, pH, and Temperature, and the Enzymatic Degradation of ASPV2 Hydrogel
2.4. ASPV2 Hydrogel as a Platform for Protein Release and Bacterial 3D Culture
2.5. Discussion
3. Conclusions
4. Materials and Methods
4.1. Bacteria and Growth Media
4.2. Plasmid Construction
4.3. Protein Expression and Purification
4.4. Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Coomassie Staining
4.5. Protein Gelation
4.6. TEM (Transmission Electron Microscopy)
4.7. ThT Assay
4.8. Bradford Assay
4.9. Hydrogel Degradation Experiment
4.10. Hydrogel Protein Releasing Test
4.11. Hydrogel-Based 3D Bacterial Culture and Recovery
4.12. Rheological Measurements
4.13. Alphafold2 Structure Modeling
4.14. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pramanik, B. Short Peptide-Based Smart Thixotropic Hydrogels. Gels 2022, 8, 569. [Google Scholar] [CrossRef]
- Correa, S.; Grosskopf, A.K.; Lopez Hernandez, H.; Chan, D.; Yu, A.C.; Stapleton, L.M.; Appel, E.A. Translational Applications of Hydrogels. Chem. Rev. 2021, 121, 11385–11457. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.E.; Hindley, J.W.; Baxani, D.K.; Ces, O.; Elani, Y. Hydrogels as Functional Components in Artificial Cell Systems. Nat. Rev. Chem. 2022, 6, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Duan, L.; Zhang, Y.; Cao, J.; Zhang, K. Current Hydrogel Advances in Physicochemical and Biological Response-Driven Biomedical Application Diversity. Signal Transduct. Target. Ther. 2021, 6, 426. [Google Scholar] [CrossRef] [PubMed]
- Khunmanee, S.; Jeong, Y.; Park, H. Crosslinking Method of Hyaluronic-Based Hydrogel for Biomedical Applications. J. Tissue Eng. 2017, 8, 204173141772646. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.C.; Lin, Q.; Xue, K.; Loh, X.J. Recent Advances in Supramolecular Hydrogels for Biomedical Applications. Mater. Today Adv. 2019, 3, 100021. [Google Scholar] [CrossRef]
- Onder, O.C.; Batool, S.R.; Nazeer, M.A. Self-Assembled Silk Fibroin Hydrogels: From Preparation to Biomedical Applications. Mater. Adv. 2022, 3, 6920–6949. [Google Scholar] [CrossRef]
- Dimatteo, R.; Darling, N.J.; Segura, T. In Situ Forming Injectable Hydrogels for Drug Delivery and Wound Repair. Adv. Drug Deliv. Rev. 2018, 127, 167–184. [Google Scholar] [CrossRef]
- Tenje, M.; Cantoni, F.; Porras Hernández, A.M.; Searle, S.S.; Johansson, S.; Barbe, L.; Antfolk, M.; Pohlit, H. A Practical Guide to Microfabrication and Patterning of Hydrogels for Biomimetic Cell Culture Scaffolds. Organs-on-a-Chip 2020, 2, 100003. [Google Scholar] [CrossRef]
- Caló, E.; Khutoryanskiy, V.V. Biomedical Applications of Hydrogels: A Review of Patents and Commercial Products. Eur. Polym. J. 2015, 65, 252–267. [Google Scholar] [CrossRef]
- Chai, Q.; Jiao, Y.; Yu, X. Hydrogels for Biomedical Applications: Their Characteristics and the Mechanisms behind Them. Gels 2017, 3, 6. [Google Scholar] [CrossRef]
- Cazzaniga, A.; Ballin, A.; Brandt, F. Hyaluronic Acid Gel Fillers in the Management of Facial Aging. Clin. Interv. Aging 2008, 3, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yue, M.; Liu, Y.; Zhang, P.; Qing, J.; Liu, H.; Zhou, Y. Advances of Engineered Hydrogel Organoids within the Stem Cell Field: A Systematic Review. Gels 2022, 8, 379. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ge, X.; Liu, L.; Xu, W.; Shao, R. Challenges and Opportunities of Silk Protein Hydrogels in Biomedical Applications. Mater. Adv. 2022, 3, 2291–2308. [Google Scholar] [CrossRef]
- Garb, J.E.; Ayoub, N.A.; Hayashi, C.Y. Untangling Spider Silk Evolution with Spidroin Terminal Domains. BMC Evol. Biol. 2010, 10, 243. [Google Scholar] [CrossRef]
- Arndt, T.; Jaudzems, K.; Shilkova, O.; Francis, J.; Johansson, M.; Laity, P.R.; Sahin, C.; Chatterjee, U.; Kronqvist, N.; Barajas-Ledesma, E.; et al. Spidroin N-Terminal Domain Forms Amyloid-like Fibril Based Hydrogels and Provides a Protein Immobilization Platform. Nat. Commun. 2022, 13, 4695. [Google Scholar] [CrossRef]
- Sivashanmugam, A.; Murray, V.; Cui, C.; Zhang, Y.; Wang, J.; Li, Q. Practical Protocols for Production of Very High Yields of Recombinant Proteins Using Escherichia coli. Protein Sci. 2009, 18, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, K.; Kono, N.; Malay, A.D.; Tateishi, A.; Ifuku, N.; Masunaga, H.; Sato, R.; Tsuchiya, K.; Ohtoshi, R.; Pedrazzoli, D.; et al. 1000 Spider Silkomes: Linking Sequences to Silk Physical Properties. Sci. Adv. 2022, 8, eabo6043. [Google Scholar] [CrossRef]
- Rising, A.; Johansson, J.; Larson, G.; Bongcam-Rudloff, E.; Engström, W.; Hjälm, G. Major Ampullate Spidroins from Euprosthenops australis: Multiplicity at Protein, mRNA and Gene Levels. Insect Mol. Biol. 2007, 16, 551–561. [Google Scholar] [CrossRef]
- Correa-Garhwal, S.M.; Babb, P.L.; Voight, B.F.; Hayashi, C.Y. Golden Orb-Weaving Spider (Trichonephila clavipes) Silk Genes with Sex-Biased Expression and Atypical Architectures. G3 Genes Genomes Genet. 2021, 11, jkaa039. [Google Scholar] [CrossRef]
- Xue, C.; Lin, T.Y.; Chang, D.; Guo, Z. Thioflavin T as an Amyloid Dye: Fibril Quantification, Optimal Concentration and Effect on Aggregation. R. Soc. Open Sci. 2017, 4, 160696. [Google Scholar] [CrossRef]
- Chan, N.J.-A.; Gu, D.; Tan, S.; Fu, Q.; Pattison, T.G.; O’Connor, A.J.; Qiao, G.G. Spider-Silk Inspired Polymeric Networks by Harnessing the Mechanical Potential of β-Sheets through Network Guided Assembly. Nat. Commun. 2020, 11, 1630. [Google Scholar] [CrossRef] [PubMed]
- Wade, R.J.; Bassin, E.J.; Rodell, C.B.; Burdick, J.A. Protease-Degradable Electrospun Fibrous Hydrogels. Nat. Commun. 2015, 6, 6639. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mooney, D.J. Designing Hydrogels for Controlled Drug Delivery. Nat. Rev. Mater. 2016, 1, 16071. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Bai, Y.; Qin, X.; Liu, J.; Huang, W.; Lv, Q. Current Understanding of Hydrogel for Drug Release and Tissue Engineering. Gels 2022, 8, 301. [Google Scholar] [CrossRef]
- Caliari, S.R.; Burdick, J.A. A Practical Guide to Hydrogels for Cell Culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Bhusari, S.; Sankaran, S.; Del Campo, A. Regulating Bacterial Behavior within Hydrogels of Tunable Viscoelasticity. Adv. Sci. 2022, 9, 2106026. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Jäkel, A.C.; Richter, J.; Eder, M.; Falgenhauer, E.; Simmel, F.C. Bacterial Growth, Communication, and Guided Chemotaxis in 3D-Bioprinted Hydrogel Environments. ACS Appl. Mater. Interfaces 2022, 14, 15871–15880. [Google Scholar] [CrossRef]
- Peerani, E.; Candido, J.B.; Loessner, D. Cell Recovery of Hydrogel-Encapsulated Cells for Molecular Analysis. In Theranostics; Batra, J., Srinivasan, S., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 2054, pp. 3–21. ISBN 978-1-4939-9768-8. [Google Scholar]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Bian, X.; Cui, C.; Qi, Y.; Sun, Y.; Zhang, Z.; Liu, W. Amino Acid Surfactant-Induced Superfast Gelation of Silk Fibroin for Treating Noncompressible Hemorrhage. Adv. Funct. Mater. 2022, 32, 2207349. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, D.; Mensaha, A.; Wang, Q.; Cai, Y.; Wei, Q. Ultrafast Gelation of Multifunctional Hydrogel/Composite Based on Self-Catalytic Fe3+/Tannic Acid-Cellulose Nanofibers. J. Colloid. Interface Sci. 2022, 606, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, J.; Magli, S.; Rabbachin, L.; Sampaolesi, S.; Nicotra, F.; Russo, L. 3D Extracellular Matrix Mimics: Fundamental Concepts and Role of Materials Chemistry to Influence Stem Cell Fate. Biomacromolecules 2020, 21, 1968–1994. [Google Scholar] [CrossRef]
- Grindy, S.C.; Learsch, R.; Mozhdehi, D.; Cheng, J.; Barrett, D.G.; Guan, Z.; Messersmith, P.B.; Holten-Andersen, N. Control of Hierarchical Polymer Mechanics with Bioinspired Metal-Coordination Dynamics. Nat. Mater. 2015, 14, 1210–1216. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Capezza, A.J.; Xiao, X.; Lendel, C.; Hedenqvist, M.S.; Kessler, V.G.; Olsson, R.T. Protein Nanofibrils and Their Hydrogel Formation with Metal Ions. ACS Nano 2021, 15, 5341–5354. [Google Scholar] [CrossRef] [PubMed]
- Park, T.G. Temperature Modulated Protein Release from pH/Temperature-Sensitive Hydrogels. Biomaterials 1999, 20, 517–521. [Google Scholar] [CrossRef]
- Sobczak, M. Enzyme-Responsive Hydrogels as Potential Drug Delivery Systems—State of Knowledge and Future Prospects. Int. J. Mol. Sci. 2022, 23, 4421. [Google Scholar] [CrossRef]
- Ashley, G.W.; Henise, J.; Reid, R.; Santi, D.V. Hydrogel Drug Delivery System with Predictable and Tunable Drug Release and Degradation Rates. Proc. Natl. Acad. Sci. USA 2013, 110, 2318–2323. [Google Scholar] [CrossRef]
- Huerta-López, C.; Alegre-Cebollada, J. Protein Hydrogels: The Swiss Army Knife for Enhanced Mechanical and Bioactive Properties of Biomaterials. Nanomaterials 2021, 11, 1656. [Google Scholar] [CrossRef]
- Xu, F.; Dawson, C.; Lamb, M.; Mueller, E.; Stefanek, E.; Akbari, M.; Hoare, T. Hydrogels for Tissue Engineering: Addressing Key Design Needs toward Clinical Translation. Front. Bioeng. Biotechnol. 2022, 10, 849831. [Google Scholar] [CrossRef]
- Gruber, D.F.; Pieribone, V.A.; Porton, B.; Kao, H.-T. Strict Regulation of Gene Expression from a High-Copy Plasmid Utilizing a Dual Vector System. Protein Expr. Purif. 2008, 60, 53–57. [Google Scholar] [CrossRef]
- Fu, S.; Wei, D.; Yang, Q.; Xie, G.; Pang, B.; Wang, Y.; Lan, R.; Wang, Q.; Dong, X.; Zhang, X.; et al. Horizontal Plasmid Transfer Promotes the Dissemination of Asian Acute Hepatopancreatic Necrosis Disease and Provides a Novel Mechanism for Genetic Exchange and Environmental Adaptation. mSystems 2020, 5, e00799-19. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, F.; Wang, Y.; Tu, B.; Cui, L. Superfast Gelation of Spider Silk-Based Artificial Silk Protein. Gels 2024, 10, 69. https://doi.org/10.3390/gels10010069
Wen F, Wang Y, Tu B, Cui L. Superfast Gelation of Spider Silk-Based Artificial Silk Protein. Gels. 2024; 10(1):69. https://doi.org/10.3390/gels10010069
Chicago/Turabian StyleWen, Fan, Yu Wang, Bowen Tu, and Lun Cui. 2024. "Superfast Gelation of Spider Silk-Based Artificial Silk Protein" Gels 10, no. 1: 69. https://doi.org/10.3390/gels10010069