Anti-Inflammatory Effects of Red Rice Bran Extract Ameliorate Type I Interferon Production via STING Pathway
Abstract
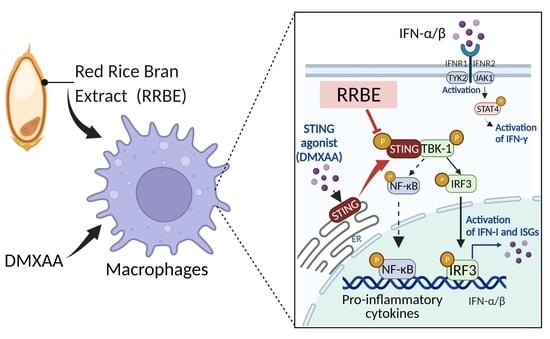
:1. Introduction
2. Materials and Methods
2.1. RRBE Preparation
2.2. Determination of the Total Flavonoid Content
2.3. Determination of the Total Phenolic Content
2.4. Determination of the Total Proanthocyanidin Content
2.5. Cell Culture
2.6. Determination of Cell Viability by the MTS Assay
2.7. Determination of Luciferase Activity
2.8. Determination of Mature Phenotypes Using Flow Cytometry
2.9. mRNA Expression Analysis
2.10. Determination of Cytokine Production
2.11. Determination of Nitric Oxide (NO) Production
2.12. Immunofluorescence
2.13. Determination of STING by Western Blot
2.14. In-Gel Digestion and Identification of Protein by Mass Spectrometry
2.15. Statistical Analysis
3. Results
3.1. Bioactive Contents of Rrice Bran Extract (RRBE)
3.2. Effects of RRBE on Cytotoxicity and the Production of Inflammatory Cytokines
3.3. Effects of RRBE on the Activation of RAW 264.7 Macrophage Cells
3.4. Anti-Inflammatory Effect of RRBE on RAW 264.7 Cells via STING Signaling
3.5. Effect of RRBE in the Activated Macrophages Decreases the Phosphorylation of STING
3.6. RRBE Promotes the Anti-Inflammatory Production of IL-10 in Macrophages
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arulselvan, P.; Fard, M.T.; Tan, W.S.; Gothai, S.; Fakurazi, S.; Norhaizan, M.E.; Kumar, S.S. Role of antioxidants and natural products in inflammation. Oxidative Med. Cell. Longev. 2016, 2016, 5276130. [Google Scholar] [CrossRef] [Green Version]
- Cronkite, D.A.; Strutt, T.M. The regulation of inflammation by innate and adaptive lymphocytes. J. Immunol. Res. 2018, 2018, 1467538. [Google Scholar] [CrossRef]
- Vincenzo, B.; Asif, I.J.; Nikolaos, P.; Francesco, M. Adaptive Immunity and Inflammation. Int. J. Inflamm. 2015, 2015, 575406. [Google Scholar] [CrossRef] [Green Version]
- Kopitar-Jerala, N. The Role of Interferons in Inflammation and Inflammasome Activation. Front. Immunol. 2017, 8, 873. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Liu, J.; Cao, X. Regulation of type I interferon signaling in immunity and inflammation: A comprehensive review. J. Autoimmun. 2017, 83, 1–11. [Google Scholar] [CrossRef]
- Muskardin, T.L.W.; Niewold, T.B. Type I interferon in rheumatic diseases. Nat. Rev. Rheumatol. 2018, 14, 214–228. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Fitzgerald-Bocarsly, P.; Feng, D. The role of type I interferon production by dendritic cells in host defense. Biochimie 2007, 89, 843–855. [Google Scholar] [CrossRef] [Green Version]
- Psarras, A.; Emery, P.; Vital, E.M. Type I interferon–mediated autoimmune diseases: Pathogenesis, diagnosis and targeted therapy. Rheumatology 2017, 56, 1662–1675. [Google Scholar] [CrossRef] [Green Version]
- Baechler, E.C.; Batliwalla, F.M.; Karypis, G.; Gaffney, P.M.; Ortmann, W.A.; Espe, K.J.; Shark, K.B.; Grande, W.J.; Hughes, K.M.; Kapur, V. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. USA 2003, 100, 2610–2615. [Google Scholar] [CrossRef] [Green Version]
- Paludan Søren, R. Activation and Regulation of DNA-Driven Immune Responses. Microbiol. Mol. Biol. Rev. 2015, 79, 225–241. [Google Scholar] [CrossRef] [Green Version]
- Shrivastav, M.; Niewold, T. Nucleic Acid Sensors and Type I Interferon Production in Systemic Lupus Erythematosus. Front. Immunol. 2013, 4, 319. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Tang, Y.-D.; Xu, G.; Zheng, C. The crosstalk between viral RNA-and DNA-sensing mechanisms. Cell. Mol. Life Sci. 2021, 78, 7427–7434. [Google Scholar] [CrossRef]
- Barber, G.N. STING: Infection, inflammation and cancer. Nat. Rev. Immunol. 2015, 15, 760–770. [Google Scholar] [CrossRef] [Green Version]
- Burdette, D.L.; Vance, R.E. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2013, 14, 19–26. [Google Scholar] [CrossRef]
- Ablasser, A.; Goldeck, M.; Cavlar, T.; Deimling, T.; Witte, G.; Röhl, I.; Hopfner, K.-P.; Ludwig, J.; Hornung, V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013, 498, 380–384. [Google Scholar] [CrossRef] [Green Version]
- Kato, K.; Omura, H.; Ishitani, R.; Nureki, O. Cyclic GMP–AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu. Rev. Biochem. 2017, 86, 541–566. [Google Scholar] [CrossRef]
- Liu, Y.; Jesus, A.A.; Marrero, B.; Yang, D.; Ramsey, S.E.; Montealegre Sanchez, G.A.; Tenbrock, K.; Wittkowski, H.; Jones, O.Y.; Kuehn, H.S. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 2014, 371, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Wei, Y.; Pandol, S.J.; Li, L.; Habtezion, A. STING Signaling Promotes Inflammation in Experimental Acute Pancreatitis. Gastroenterology 2018, 154, 1822–1835.e2. [Google Scholar] [CrossRef] [Green Version]
- Baiges, I.; Palmfeldt, J.; Blade, C.; Gregersen, N.; Arola, L. Lipogenesis is decreased by grape seed proanthocyanidins according to liver proteomics of rats fed a high fat diet. Mol. Cell. Proteom. 2010, 9, 1499–1513. [Google Scholar] [CrossRef] [Green Version]
- Karlsen, A.; Retterstøl, L.; Laake, P.; Paur, I.; Kjølsrud-Bøhn, S.; Sandvik, L.; Blomhoff, R. Anthocyanins inhibit nuclear factor-κ B activation in monocytes and reduce plasma concentrations of pro-inflammatory mediators in healthy adults. J. Nutr. 2007, 137, 1951–1954. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Li, Y. Grape seed proanthocyanidin extracts prevent hyperglycemia-induced monocyte adhesion to aortic endothelial cells and ameliorates vascular inflammation in high-carbohydrate/high-fat diet and streptozotocin-induced diabetic rats. Int. J. Food Sci. Nutr. 2016, 67, 524–534. [Google Scholar] [CrossRef]
- Aboonabi, A.; Aboonabi, A. Anthocyanins reduce inflammation and improve glucose and lipid metabolism associated with inhibiting nuclear factor-kappaB activation and increasing PPAR-γ gene expression in metabolic syndrome subjects. Free Radic. Biol. Med. 2020, 150, 30–39. [Google Scholar] [CrossRef]
- Boue, S.M.; Daigle, K.W.; Chen, M.-H.; Cao, H.; Heiman, M.L. Antidiabetic Potential of Purple and Red Rice (Oryza sativa L.) Bran Extracts. J. Agric. Food Chem. 2016, 64, 5345–5353. [Google Scholar] [CrossRef]
- Surarit, W.; Jansom, C.; Lerdvuthisopon, N.; Kongkham, S.; Hansakul, P. Evaluation of antioxidant activities and phenolic subtype contents of ethanolic bran extracts of Thai pigmented rice varieties through chemical and cellular assays. Int. J. Food Sci. Technol. 2015, 50, 990–998. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Lai, H.-M. Bioactive compounds and antioxidative activity of colored rice bran. J. Food Drug Anal. 2016, 24, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.W.; Kobayashi, K.; Shen, L.; Inagaki, J.; Ide, M.; Hwang, S.S.; Matsuura, E. Antioxidative attributes of rice bran extracts in ameliorative effects of atherosclerosis-associated risk factors. Heliyon 2020, 6, e05743. [Google Scholar] [CrossRef]
- Limtrakul, P.; Yodkeeree, S.; Pitchakarn, P.; Punfa, W. Anti-inflammatory effects of proanthocyanidin-rich red rice extract via suppression of MAPK, AP-1 and NF-κB pathways in Raw 264.7 macrophages. Nutr. Res. Pract. 2016, 10, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Thim-Uam, A.; Prabakaran, T.; Tansakul, M.; Makjaroen, J.; Wongkongkathep, P.; Chantaravisoot, N.; Saethang, T.; Leelahavanichkul, A.; Benjachat, T.; Paludan, S.R. STING mediates lupus via the activation of conventional dendritic cell maturation and plasmacytoid dendritic cell differentiation. Iscience 2020, 23, 101530. [Google Scholar] [CrossRef]
- Papinska, J.; Bagavant, H.; Gmyrek, G.B.; Sroka, M.; Tummala, S.; Fitzgerald, K.A.; Deshmukh, U.S. Activation of Stimulator of Interferon Genes (STING) and Sjögren Syndrome. J. Dent. Res. 2018, 97, 893–900. [Google Scholar] [CrossRef]
- Xiong, H.; Xi, Y.; Yuan, Z.; Wang, B.; Hu, S.; Fang, C.; Cai, Y.; Fu, X.; Li, L. IFN-γ activates the tumor cell-intrinsic STING pathway through the induction of DNA damage and cytosolic dsDNA formation. OncoImmunology 2022, 11, 2044103. [Google Scholar] [CrossRef]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef]
- Couillin, I.; Riteau, N. STING Signaling and Sterile Inflammation. Front. Immunol. 2021, 12, 753789. [Google Scholar] [CrossRef]
- Ghasemzadeh, A.; Karbalaii, M.T.; Jaafar, H.Z.E.; Rahmat, A. Phytochemical constituents, antioxidant activity, and antiproliferative properties of black, red, and brown rice bran. Chem. Cent. J. 2018, 12, 17. [Google Scholar] [CrossRef] [Green Version]
- Ciulu, M.; Cádiz-Gurrea, M.D.; Segura-Carretero, A. Extraction and Analysis of Phenolic Compounds in Rice: A Review. Molecules 2018, 23, 2890. [Google Scholar] [CrossRef] [Green Version]
- Qi, Q.; Chu, M.; Yu, X.; Xie, Y.; Li, Y.; Du, Y.; Liu, X.; Zhang, Z.; Shi, J.; Yan, N. Anthocyanins and Proanthocyanidins: Chemical Structures, Food Sources, Bioactivities, and Product Development. Food Rev. Int. 2022, 38, 1–29. [Google Scholar] [CrossRef]
- Pattananandecha, T.; Apichai, S.; Sirilun, S.; Julsrigival, J.; Sawangrat, K.; Ogata, F.; Kawasaki, N.; Sirithunyalug, B.; Saenjum, C. Anthocyanin Profile, Antioxidant, Anti-Inflammatory, and Antimicrobial against Foodborne Pathogens Activities of Purple Rice Cultivars in Northern Thailand. Molecules 2021, 26, 5234. [Google Scholar] [CrossRef]
- Yazhen, S.; Wenju, W.; Panpan, Z.; Yuanyuan, Y.; Panpan, D.; Wusen, Z.; Yanling, W. Anthocyanins: Novel antioxidants in diseases prevention and human health. In Flavonoids-A Coloring Model for Cheering Up Life; IntechOpen: London, UK, 2019; pp. 1–16. [Google Scholar]
- Junmarkho, K.; Hansakul, P. Thai pigmented rice bran extracts inhibit production of superoxide, nitric oxide radicals and inducible nitric oxide synthase in cellular models. Asian Pac. J. Trop. Biomed. 2019, 9, 291. [Google Scholar]
- Saji, N.; Schwarz, L.J.; Santhakumar, A.B.; Blanchard, C.L. Stabilization treatment of rice bran alters phenolic content and antioxidant activity. Cereal Chem. 2020, 97, 281–292. [Google Scholar] [CrossRef]
- Saji, N.; Francis, N.; Schwarz, L.J.; Blanchard, C.L.; Santhakumar, A.B. The Antioxidant and Anti-Inflammatory Properties of Rice Bran Phenolic Extracts. Foods 2020, 9, 829. [Google Scholar] [CrossRef]
- Buxadé, M.; Huerga Encabo, H.; Riera-Borrull, M.; Quintana-Gallardo, L.; López-Cotarelo, P.; Tellechea, M.; Martínez-Martínez, S.; Redondo, J.M.; Martín-Caballero, J.; Flores, J.M.; et al. Macrophage-specific MHCII expression is regulated by a remote Ciita enhancer controlled by NFAT5. J. Exp. Med. 2018, 215, 2901–2918. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.; Ashkar, A. The dual nature of type I and type II interferons. Front Immunol. 2018, 9, 2061. [Google Scholar] [CrossRef] [Green Version]
- Michlmayr, D.; McKimmie, C.S. Role of CXCL10 in central nervous system inflammation. Int. J. Interferon Cytokine Mediat. Res. 2014, 6, 1. [Google Scholar]
- Kota, R.S.; Rutledge, J.C.; Gohil, K.; Kumar, A.; Enelow, R.I.; Ramana, C.V. Regulation of gene expression in RAW 264.7 macrophage cell line by interferon-γ. Biochem. Biophys. Res. Commun. 2006, 342, 1137–1146. [Google Scholar] [CrossRef]
- Rashighi, M.; Harris, J.E. Interfering with the IFN-γ/CXCL10 pathway to develop new targeted treatments for vitiligo. Ann. Transl. Med. 2015, 3, 343. [Google Scholar]
- Chan, E.D.; Riches, D.W. IFN-γ+ LPS induction of iNOS is modulated by ERK, JNK/SAPK, and p38 mapk in a mouse macrophage cell line. Am. J. Physiol.-Cell Physiol. 2001, 280, C441–C450. [Google Scholar] [CrossRef]
- Chen, C.-W.; Chang, Y.-H.; Tsi, C.-J.; Lin, W.-W. Inhibition of IFN-γ-Mediated Inducible Nitric Oxide Synthase Induction by the Peroxisome Proliferator-Activated Receptor γ Agonist, 15-Deoxy-Δ12,14-Prostaglandin J2, Involves Inhibition of the Upstream Janus Kinase/STAT1 Signaling Pathway. J. Immunol. 2003, 171, 979. [Google Scholar] [CrossRef] [Green Version]
- Blanchette, J.; Jaramillo, M.; Olivier, M. Signalling events involved in interferon-γ-inducible macrophage nitric oxide generation. Immunology 2003, 108, 513–522. [Google Scholar] [CrossRef]
- Mishra, B.B.; Rathinam, V.A.K.; Martens, G.W.; Martinot, A.J.; Kornfeld, H.; Fitzgerald, K.A.; Sassetti, C.M. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome–dependent processing of IL-1β. Nat. Immunol. 2013, 14, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wilson, H.L.; Kiss-Toth, E. Regulating STING in health and disease. J. Inflamm. 2017, 14, 11. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira Mann, C.C.; Orzalli, M.H.; King, D.S.; Kagan, J.C.; Lee, A.S.Y.; Kranzusch, P.J. Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-κB Signaling Adaptation. Cell Rep. 2019, 27, 1165–1175.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Z.; Ma, T.; Liu, X.; Wang, C. cGAS-STING pathway: Post-translational modifications and functions in sterile inflammatory diseases. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hopfner, K.-P.; Hornung, V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 2020, 21, 501–521. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Chen, Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012, 5, ra20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarassishin, L.; Suh, H.-S.; Lee, S.C. Interferon regulatory factor 3 plays an anti-inflammatory role in microglia by activating the PI3K/Akt pathway. J. Neuroinflammation 2011, 8, 187. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Barber, G.N. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 2014, 88, 5328–5341. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Dai, T.; He, X.; Zhang, Z.; Xie, F.; Wang, S.; Zhang, L.; Zhou, F. The interactions between cGAS-STING pathway and pathogens. Signal Transduct. Target. Ther. 2020, 5, 91. [Google Scholar] [CrossRef]
- Burdette, D.L.; Monroe, K.M.; Sotelo-Troha, K.; Iwig, J.S.; Eckert, B.; Hyodo, M.; Hayakawa, Y.; Vance, R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011, 478, 515–518. [Google Scholar] [CrossRef]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.-Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; et al. The Adaptor Protein MITA Links Virus-Sensing Receptors to IRF3 Transcription Factor Activation. Immunity 2008, 29, 538–550. [Google Scholar] [CrossRef] [Green Version]
- Iyer, S.S.; Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef] [Green Version]
- Benveniste, E.N.; Qin, H. Type I interferons as anti-inflammatory mediators. Sci. STKE 2007, 2007, pe70. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Son, S.; Oliveira, S.C.; Barber, G.N. STING-dependent signaling underlies IL-10 controlled inflammatory colitis. Cell Rep. 2017, 21, 3873–3884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, B.M.; Uchimura, T.; Gallovic, M.D.; Thamilarasan, M.; Chou, W.-C.; Gibson, S.A.; Deng, M.; Tam, J.W.; Batty, C.J.; Williams, J. STING Agonist Mitigates Experimental Autoimmune Encephalomyelitis by Stimulating Type I IFN–Dependent and–Independent Immune-Regulatory Pathways. J. Immunol. 2021, 206, 2015–2028. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Rutz, S.; Crellin, N.K.; Valdez, P.A.; Hymowitz, S.G. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 2011, 29, 71–109. [Google Scholar] [CrossRef]







| Bioactive | Constituents |
|---|---|
| Total phenolic content (mg GAE/g) | 51.9 ± 1.73 |
| Total flavonoid content (mg Catechin/g) | 22.94 ± 2.62 |
| Proanthocyanidins (mg Catechin/g) | 6.52 ± 0.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onsa-Ard, A.; Thongboontho, R.; Munkong, N.; Phromnoi, K.; Ontawong, A.; Pengnet, S.; Thim-Uam, A. Anti-Inflammatory Effects of Red Rice Bran Extract Ameliorate Type I Interferon Production via STING Pathway. Foods 2022, 11, 1622. https://doi.org/10.3390/foods11111622
Onsa-Ard A, Thongboontho R, Munkong N, Phromnoi K, Ontawong A, Pengnet S, Thim-Uam A. Anti-Inflammatory Effects of Red Rice Bran Extract Ameliorate Type I Interferon Production via STING Pathway. Foods. 2022; 11(11):1622. https://doi.org/10.3390/foods11111622
Chicago/Turabian StyleOnsa-Ard, Amnart, Rungthip Thongboontho, Narongsuk Munkong, Kanokkarn Phromnoi, Atcharaporn Ontawong, Sirinat Pengnet, and Arthid Thim-Uam. 2022. "Anti-Inflammatory Effects of Red Rice Bran Extract Ameliorate Type I Interferon Production via STING Pathway" Foods 11, no. 11: 1622. https://doi.org/10.3390/foods11111622