Genome-Wide Identification and Transcriptional Expression Profiles of the F-box Gene Family in Common Walnut (Juglans regia L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis of Putative F-box Proteins from J. regia
2.2. F-box Protein Alignment, Phylogenetic Analysis, Pfam Domain Detection, and Chromosome Location Analysis of Common Walnut F-box Genes
2.3. The KEGG Pathway, Promoter, Gene Structure, and Motif Analysis of J. regia F-box Genes
2.4. Synteny Analysis, and Calculating Ka, Ks, and Ka/Ks Values of Duplicated Gene Pairs
2.5. Microarray Expression Profiles of F-box Genes
2.6. Plant Materials, Treatments, and Collections
3. Results
3.1. Identification, Classification, and Genomic Distribution of F-box Genes in J. regia
3.2. Phylogenetic Relationship, Gene Structure, and Protein Domains of the F-box Gene Family of Common Walnut
3.3. KEGG Analysis, and Cis-Acting Regulatory Elements in the Promoter of the F-box Gene Family of Common Walnut
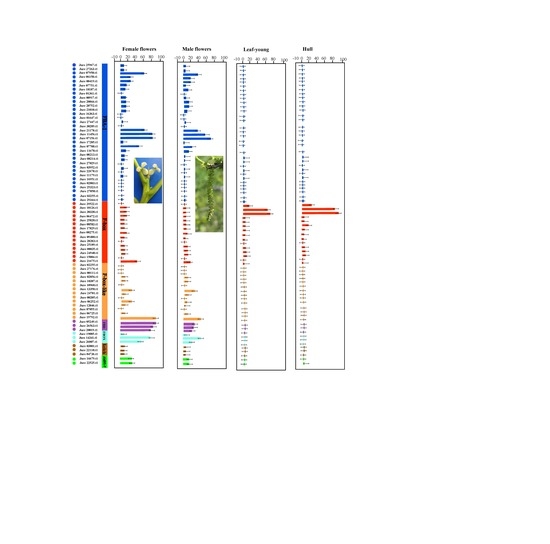
3.4. Expression Profile Analysis of Walnut F-box Genes
4. Discussion
4.1. The Characteristics of the F-box Gene Family of Common Walnut
4.2. The Expansion and Evolution of the F-box Gene Family of Common Walnut
4.3. Function of the F-box Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kumar, A.; Paietta, J.V. The sulfur controller-2 negative regulatory gene of Neurospora crassa encodes a protein with beta-transducin repeats. Proc. Natl. Acad. Sci. USA 1995, 92, 3343–3347. [Google Scholar] [CrossRef] [PubMed]
- Kipreos, E.T.; Pagano, M. The F-box protein family. Genome Biol. 2000, 1, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Ang, X.L.; Shirogane, T.; Harper, J.W. Identification of substrates for F-box proteins. Method Enzym. 2005, 399, 287–309. [Google Scholar]
- Xu, G.X.; Hong, M.; Masatoshi, N.; Hongzhi, K. Evolution of F-box genes in plants: Different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. USA 2009, 106, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Stefanowicz, K.; Lannoo, N.; Zhao, Y.; Eggermont, L.; Hove, J.V.; Atalah, B.A.; Damme, E.J.M.V. Glycan-binding F-box protein from Arabidopsis thaliana protects plants from pseudomonas syringae infection. BMC Plant Biol. 2016, 16, 213. [Google Scholar] [CrossRef] [PubMed]
- Baute, J.; Polyn, S.; De, B.J.; Blomme, J.; Van, L.M.; Inzé, D. F-box protein FBX92 affects leaf size in Arabidopsis thaliana. Plant Cell Physiol. 2017, 58, 962–975. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, G.; Zhou, S.; Ren, Y.; Wang, W. The improvement of salt tolerance in transgenic tobacco by overexpression of wheat F-box gene TaFBA1. Plant Sci. 2017, 259, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Cooley, L. Kelch encodes a component of intercellular bridges in drosophila egg chambers. Cell 1993, 72, 681–693. [Google Scholar] [CrossRef]
- Woo, H.R.; Chung, K.M.; Park, J.H.; Oh, S.A.; Ahn, T.; Hong, S.H.; Jang, S.K.; Nam, H.G. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 2001, 13, 1779–1790. [Google Scholar] [CrossRef]
- Petra, S.; Furner, I.J.; Ottoline Leyser, H.M. Max2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007, 50, 80–94. [Google Scholar]
- Durfee, T.; Roe, J.L.; Sessions, R.A.; Inouye, C.; Serikawa, K.; Feldmann, K.A.; Weigel, D.; Zambryski, P.C. The F-box-containing protein ufo and agamous participate in antagonistic pathways governing early petal development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 8571–8576. [Google Scholar] [CrossRef] [PubMed]
- Takato, I.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of constans in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar]
- Eva, K.; Péter, G.; Anthony, H.; László, K.B.; Woe-Yeon, K.; Eriksson, M.E.; Réka, T.; Shigeru, H.; Balázs, F.; Southern, M.M. Forward genetic analysis of the circadian clock separates the multiple functions of zeitlupe. Plant Physiol. 2006, 140, 933–945. [Google Scholar]
- Han, L.Q.; Mason, M.E.; Risseeuw, E.P.; Crosby, W.L.; Somers, D.E. Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J. 2010, 40, 291–301. [Google Scholar] [CrossRef]
- Paja, S.; Xi, W.; Skirpan, A.L.; Yan, W.; Dowd, P.E.; Mccubbin, A.G.; Shihshieh, H.; Teh-Hui, K. Identification of the pollen determinant of s-RNase-mediated self-incompatibility. Nature 2004, 429, 302–305. [Google Scholar]
- Hong, Q.; Fei, W.; Lan, Z.; Zhou, J.; Zhao, L.; Zhang, Y.; Robbins, T.P.; Xue, Y. The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 2004, 16, 2307–2322. [Google Scholar]
- Wang, L.; Dong, L.; Zhang, Y.E.; Zhang, Y.; Wu, W.; Deng, X.; Xue, Y. Genome-wide analysis of s-locus F-box-like genes in Arabidopsis thaliana. Plant Mol. Biol. 2004, 56, 929–945. [Google Scholar] [CrossRef]
- Dieterle, M.; Zhou, Y.C.; SchäFer, E.; Funk, M.; Kretsch, T. Eid1, an F-box protein involved in phytochrome a-specific light signaling. Genes Dev. 2001, 15, 939–944. [Google Scholar] [CrossRef]
- Alyssa, D.; Thomas, S.G.; Jianhong, H.; Steber, C.M.; Tai-Ping, S. The Arabidopsis F-box protein sleepy1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 2004, 16, 1392–1405. [Google Scholar]
- Binder, B.M.; Walker, J.M.; Gagne, J.M.; Emborg, T.J.; Georg, H.; Bleecker, A.B.; Vierstra, R.D. The arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 2007, 19, 509–523. [Google Scholar] [CrossRef]
- Calderón-Villalobos, L.I.A.; Nill, C.; Marrocco, K.; Kretsch, T.; Schwechheimer, C. The evolutionarily conserved arabidopsis thaliana F-box protein AtFBP7 is required for efficient translation during temperature stress. Gene 2007, 392, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Suk, K.H.; Delaney, T.P. Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 2002, 14, 1469–1482. [Google Scholar]
- Gagne, J.M.; Downes, B.P.; Shin-Han, S.; Durski, A.M.; Vierstra, R.D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.R.; Zhang, Z.R.; Lv, W.; Xu, J.N.; Wang, X.Y. Genome-wide characterization and analysis of F-box protein-encoding genes in the Malus domestica genome. Mol. Genet. Genom. 2015, 290, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Mukesh, J.; Aashima, N.; Rita, A.; Pinky, A.; Swatismita, R.; Pooja, S.; Sanjay, K.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar]
- Jia, F.J.; Wu, B.J.; Li, H.; Huang, J.G.; Zheng, C.C. Genome-wide identification and characterisation of F-box family in maize. Mol. Genet. Genom. 2013, 288, 559–577. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Xiao, Z.X.; Wong, F.L.; Sun, S.; Liang, K.J.; Lam, H.M. Genome-wide analyses of the soybean F-box gene family in response to salt stress. Int. J. Mol. Sci. 2017, 18, 818. [Google Scholar] [CrossRef]
- Gupta, S.; Garg, V.; Kant, C.; Bhatia, S. Genome-wide survey and expression analysis of F-box genes in chickpea. BMC Genom. 2015, 16, 67. [Google Scholar] [CrossRef]
- Wang, G.M.; Yin, H.; Qiao, X.; Tan, X.; Gu, C.; Wang, B.H.; Cheng, R.; Wang, Y.Z.; Zhang, S.L. F-box genes: Genome-wide expansion, evolution and their contribution to pollen growth in pear (pyrus bretschneideri). Plant Sci. 2016, 253, 164–175. [Google Scholar] [CrossRef]
- Yang, X.H.; Kalluri, U.C.; Sara, J.; Gunter, L.E.; Tongming, Y.; Tschaplinski, T.J.; Weston, D.J.; Priya, R.; Tuskan, G.A. The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 2008, 148, 1189–1200. [Google Scholar] [CrossRef]
- Jian, B.S.; Yan, X.W.; Hai, B.L.; Bo, W.L.; Zhao, S.Z.; Shuai, G.; Zhi, M.Y. The F-box family genes as key elements in response to salt, heavy mental, and drought stresses in Medicago truncatula. Funct. Integr. Genom. 2015, 15, 495–507. [Google Scholar]
- Hidenori, S.; Hiroyuki, K.; Mayu, M.; Yusuke, S.; Toshio, H.; Koichiro, U.; Makoto, K.; Hisashi, H.; Takato, K.J.G. S locus F-box brothers: Multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 2007, 175, 1869–1881. [Google Scholar]
- Ushijima, K.; Yamane, H.; Watari, A.; Kakehi, E.; Ikeda, K.; Hauck, N.R.; Iezzoni, A.F.; Tao, R.J.P.J. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 2010, 39, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Entani, T.; Iwano, M.; Shiba, H.; Che, F.S.; Isogai, A.; Takayama, S.J.G.t.C. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: Identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 2010, 8, 203–213. [Google Scholar] [CrossRef]
- Ikeda, K.; Igic, B.; Ushijima, K.; Yamane, H.; Hauck, N.R.; Nakano, R.; Sassa, H.; Iezzoni, A.F.; Kohn, J.R.; Tao, R.J.S.P.R. Primary structural features of the S haplotype-specific F-box protein, SFB, in Prunus. Sex. Plant Reprod. 2004, 16, 235–243. [Google Scholar] [CrossRef]
- Hiroyuki, K.; Masaki, K.; Koichiro, U.; Miyoko, K.; Shu, K.; Hidenori, S.J.P.J. Sequence divergence and loss-of-function phenotypes of S locus F-box brothers genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia). Plant J. 2011, 68, 1028–1038. [Google Scholar]
- Han, H.; Woeste, K.E.; Hu, Y.; Meng, D.; Tian, Z.; Gao, X.X.; Zhou, H.; Feng, X.; Zhao, G.; Peng, Z. Genetic diversity and population structure of common walnut (Juglans regia) in china based on est-ssrs and the nuclear gene phenylalanine ammonia-lyase (PAL). Tree Genet. Genomes 2016, 12, 111. [Google Scholar] [CrossRef]
- Hu, Y.; Woeste, K.E.; Zhao, P. Completion of the chloroplast genomes of five chinese juglans and their contribution to chloroplast phylogeny. Front. Plant Sci. 2016, 7, 1955. [Google Scholar] [CrossRef]
- Zhao, P.; Zhou, H.J.; Potter, D.; Hu, Y.H.; Feng, X.J.; Dang, M.; Feng, L.; Zulfiqar, S.; Liu, W.Z.; Zhao, G. Population genetics, phylogenomics and hybrid speciation of juglans in china determined from whole chloroplast genomes, transcriptomes, and genotyping-by-sequencing (GBS). Mol. Phylogenet. Evol. 2018, 126, 250–265. [Google Scholar] [CrossRef]
- Feng, X.; Yuan, X.; Sun, Y.; Hu, Y.; Zulfiqar, S.; Ouyang, X.; Meng, D.; Zhou, H.; Woeste, K.; Peng, Z. Resources for studies of iron walnut (Juglans sigillata) gene expression, genetic diversity, and evolution. Tree Genet. Genomes 2018, 14, 51. [Google Scholar] [CrossRef]
- Martínez-García, P.J.; Crepeau, M.W.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of nonstructural polyphenols. Plant J. 2016, 87, 507–532. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. Mega: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Yan, F.; Li, H.Z.; Zhao, P. Genome-Wide Identification and transcriptional expression of the PAL Gene family in common Walnut (Juglans regia L.). Genes 2019, 10, 46. [Google Scholar] [CrossRef]
- Magali, L.; Patrice, D.; Gert, T.; Kathleen, M.; Yves, M.; Yves, V.D.P.; Pierre, R.; Stephane, R. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The meme suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H.; et al. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Rozas, J.; Librado, P. Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar]
- Doerks, T.; Copley, R.R.; Schultz, J.; Ponting, C.P.; Bork, P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002, 12, 47–56. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Deng, W.K.; Wang, Y.B.; Liu, Z.X.; Chen, H.; Xue, Y. Hemi: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- Zhu, H.; Han, X.; Lv, J.; Zhao, L.; Xu, X.; Zhang, T.; Guo, W. Structure, expression differentiation and evolution of duplicated fiber developmental genes in gossypium barbadense and G. hirsutum. BMC Plant Biol. 2011, 11, 40. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Moran, Y.; Levin, J.Z.; Thompson, D.A.; Ido, A.; Xian, A.; Lin, F.; Raktima, R.; Qiandong, Z. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Li, P.; Piao, Y.; Shon, H.S.; Ryu, K.H. Comparing the normalization methods for the differential analysis of illumina high-throughput RNA-seq data. BMC Bioinform. 2015, 16, 347. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Ana, C.; Stefan, G.T.; Juan Miguel, G.G.; Javier, T.; Manuel, T.; Montserrat, R. Blast2go: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Marienfeld, J.; Unseld, M.; Brandt, P.; Brennicke, A. Genomic recombination of the mitochondrial Atp6 gene in Arabidopsis thaliana at the protein processing site creates two different presequences. DNA Res. 1996, 3, 287–290. [Google Scholar] [CrossRef]
- Goff, S.A.; Darrell, R.; Tien-Hung, L.; Gernot, P.; Ronglin, W.; Molly, D.; Jane, G.; Allen, S.; Paul, O.; Hemant, V. A draft sequence of the rice genome (Oryza sativa L; Ssp. Japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef]
- Hua, Z.H.; Cheng, Z.; Shin-Han, S.; Vierstra, R.D. Phylogenetic comparison of F-box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 2011, 6, e16219. [Google Scholar] [CrossRef]
- Initiative, A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Li, W.; Xie, X.; Lu, Y.; Liu, Y.; Jin, X.; Suo, Z. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Front. Plant Sci. 2017, 8, 1148. [Google Scholar] [CrossRef]
- Gorlova, O.; Fedorov, A.; Logothetis, C.; Amos, C.; Gorlov, I. Genes with a large intronic burden show greater evolutionary conservation on the protein level. BMC Evol. Biol. 2014, 14, 50. [Google Scholar] [CrossRef]
- Long, M.; Deutsch, M. Association of intron phases with conservation at splice site sequences and evolution of spliceosomal introns. Mol. Biol. Evol. 1999, 16, 1528–1534. [Google Scholar] [CrossRef] [Green Version]
- Zheng, T.Z.; Zhuo, X.K.; Li, L.L.; Wang, J.; Cheng, T.R.; Zhang, Q.X. Genome-Wide analysis of the D-type cyclin gene family reveals differential expression patterns and stem development in the woody plant Prunus mume. Forests 2019, 10, 147. [Google Scholar] [CrossRef]
- Islam, M.S.; Choudhury, M.; Majlish, A.N.K.; Islam, T.; Ghosh, A. Comprehensive genome-wide analysis of glutathione s-transferase gene family in potato (Solanum tuberosum L.) and their expression profiling in various anatomical tissues and perturbation conditions. Gene 2017, 639, 149–162. [Google Scholar] [CrossRef]
- Dong, C.-J.; Shang, Q.-M. Genome-wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta 2013, 238, 35–49. [Google Scholar] [CrossRef]
- Dinant, S.; Clark, A.M.; Zhu, Y.; Vilaine, F.; Palauqui, J.C.; Kusiak, C.; Thompson, G.A.; Thompson, G.A. Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol. 2003, 131, 114–128. [Google Scholar] [CrossRef]
- Hofmann, K. Skp1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996, 86, 263–274. [Google Scholar]
- Wang, N.Y.; Shirogane, T.; Liu, D.; Harper, J.W.; Elledge, S.J. Exit from exit: Resetting the cell cycle through am inhibition of g protein signaling. Cell 2003, 112, 697–709. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, F.; Zhou, H.; Yue, M.; Yang, G.; Li, H.; Zhang, S.; Zhao, P. Genome-Wide Identification and Transcriptional Expression Profiles of the F-box Gene Family in Common Walnut (Juglans regia L.). Forests 2019, 10, 275. https://doi.org/10.3390/f10030275
Yan F, Zhou H, Yue M, Yang G, Li H, Zhang S, Zhao P. Genome-Wide Identification and Transcriptional Expression Profiles of the F-box Gene Family in Common Walnut (Juglans regia L.). Forests. 2019; 10(3):275. https://doi.org/10.3390/f10030275
Chicago/Turabian StyleYan, Feng, Huijuan Zhou, Ming Yue, Ge Yang, Huaizhu Li, Shuoxin Zhang, and Peng Zhao. 2019. "Genome-Wide Identification and Transcriptional Expression Profiles of the F-box Gene Family in Common Walnut (Juglans regia L.)" Forests 10, no. 3: 275. https://doi.org/10.3390/f10030275