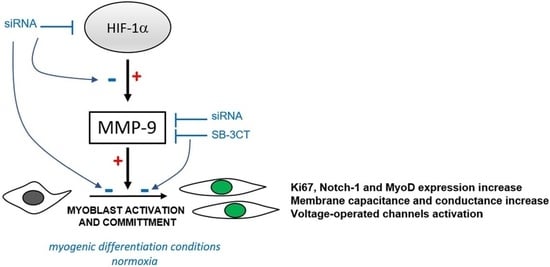
HIF-1α/MMP-9 Axis Is Required in the Early Phases of Skeletal Myoblast Differentiation under Normoxia Condition In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Silencing of HIF-1a and MMP-9 by Short Interfering RNA
2.3. Morphological Analyses
2.3.1. Phase Contrast Microscopy
2.3.2. Confocal Laser Scanning Microscopy
2.4. Total RNA Extraction and Semi Quantitative Reverse Transcription (RT)-PCR
2.5. Western Blotting
2.6. Electrophysiological Recordings
2.7. Statistical Analysis
3. Results
3.1. HIF-1α and MMP-9 Expression during C2C12 Myoblast Differentiation under Normoxia
3.2. HIF-1α and MMP-9 Are Required for Myogenic Commitment of Myoblasts and MMP-9 Is an Effector of HIF-1α
3.2.1. Morphological and Biochemical Analyses
3.2.2. Functional Electrophysiological Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sousa-Victor, P.; García-Prat, L.; Muñoz-Cánoves, P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 2022, 23, 204–226. [Google Scholar] [CrossRef] [PubMed]
- Feige, P.; Brun, C.E.; Ritso, M.; Rudnicki, M.A. Orienting Muscle Stem Cells for Regeneration in Homeostasis, Aging, and Disease. Cell Stem Cell 2018, 23, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L.; Kamal, M.; Parise, G. The Role of Supporting Cell Populations in Satellite Cell Mediated Muscle Repair. Cells 2023, 12, 1968. [Google Scholar] [CrossRef] [PubMed]
- Forcina, L.; Cosentino, M.; Musarò, A. Mechanisms Regulating Muscle Regeneration: Insights into the Interrelated and Time-Dependent Phases of Tissue Healing. Cells 2020, 9, 1297. [Google Scholar] [CrossRef] [PubMed]
- Manetti, M.; Tani, A.; Rosa, I.; Chellini, F.; Squecco, R.; Idrizaj, E.; Zecchi-Orlandini, S.; Ibba-Manneschi, L.; Sassoli, C. Morphological evidence for telocytes as stromal cells supporting satellite cell activation in eccentric contraction-induced skeletal muscle injury. Sci. Rep. 2019, 9, 14515. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.A.; Olsen, I.; Zammit, P.S.; Heslop, L.; Petrie, A.; Partridge, T.A.; Morgan, J.E. Stem Cell Function, Self-Renewal, and Behavioral Heterogeneity of Cells from the Adult Muscle Satellite Cell Niche. Cell 2005, 122, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Takemasa, T.; Fujita, R. High Throughput Screening of Mitochondrial Bioenergetics in Myoblasts and Differentiated Myotubes. Methods Mol. Biol. 2023, 2640, 89–98. [Google Scholar] [CrossRef]
- Singh, V. Intracellular metabolic reprogramming mediated by micro-RNAs in differentiating and proliferating cells under non-diseased conditions. Mol. Biol. Rep. 2021, 48, 8123–8140. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Scimè, A. Mitochondrial Function in Muscle Stem Cell Fates. Front. Cell Dev. Biol. 2020, 8, 480. [Google Scholar] [CrossRef]
- Cirillo, F.; Mangiavini, L.; La Rocca, P.; Piccoli, M.; Ghiroldi, A.; Rota, P.; Tarantino, A.; Canciani, B.; Coviello, S.; Messina, C.; et al. Human Sarcopenic Myoblasts Can Be Rescued by Pharmacological Reactivation of HIF-1α. Int. J. Mol. Sci. 2022, 23, 7114. [Google Scholar] [CrossRef]
- Salekeen, R.; Kyba, M. Not young but still immature: A HIF-1α-mediated maturation checkpoint in regenerating muscle. J. Clin. Investig. 2022, 132, e165322. [Google Scholar] [CrossRef]
- Lu, Y.; Mao, J.; Han, X.; Zhang, W.; Li, Y.; Liu, Y.; Li, Q. Downregulated hypoxia-inducible factor 1α improves myoblast differentiation under hypoxic condition in mouse genioglossus. Mol. Cell. Biochem. 2021, 476, 1351–1364. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Conotte, S.; Belayew, A.; Declèves, A.E.; Legrand, A.; Tassin, A. Hypoxia and Hypoxia-Inducible Factor Signaling in Muscular Dystrophies: Cause and Consequences. Int. J. Mol. Sci. 2021, 22, 7220. [Google Scholar] [CrossRef]
- Pircher, T.; Wackerhage, H.; Aszodi, A.; Kammerlander, C.; Böcker, W.; Saller, M.M. Hypoxic Signaling in Skeletal Muscle Maintenance and Regeneration: A Systematic Review. Front. Physiol. 2021, 12, 684899. [Google Scholar] [CrossRef]
- Cirillo, F.; Resmini, G.; Angelino, E.; Ferrara, M.; Tarantino, A.; Piccoli, M.; Rota, P.; Ghiroldi, A.; Monasky, M.M.; Ciconte, G.; et al. HIF-1α Directly Controls WNT7A Expression During Myogenesis. Front. Cell Dev. Biol. 2020, 8, 593508. [Google Scholar] [CrossRef]
- Sinha, K.M.; Tseng, C.; Guo, P.; Lu, A.; Pan, H.; Gao, X.; Andrews, R.; Eltzschig, H.; Huard, J. Hypoxia-inducible factor 1α (HIF-1α) is a major determinant in the enhanced function of muscle-derived progenitors from MRL/MpJ mice. FASEB J. 2019, 33, 8321–8334. [Google Scholar] [CrossRef]
- Cirillo, F.; Resmini, G.; Ghiroldi, A.; Piccoli, M.; Bergante, S.; Tettamanti, G.; Anastasia, L. Activation of the hypoxia-inducible factor 1α promotes myogenesis through the noncanonical Wnt pathway, leading to hypertrophic myotubes. FASEB J. 2017, 31, 2146–2156. [Google Scholar] [CrossRef]
- Yang, X.; Yang, S.; Wang, C.; Kuang, S. The hypoxia-inducible factors HIF1α and HIF2α are dispensable for embryonic muscle development but essential for postnatal muscle regeneration. J. Biol. Chem. 2017, 292, 5981–5991. [Google Scholar] [CrossRef] [PubMed]
- Dehne, N.; Kerkweg, U.; Otto, T.; Fandrey, J. The HIF-1 response to simulated ischemia in mouse skeletal muscle cells neither enhances glycolysis nor prevents myotube cell death. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 293, R1693–R1701. [Google Scholar] [CrossRef]
- Li, X.; Zhu, L.; Chen, X.; Fan, M. Effects of hypoxia on proliferation and differentiation of myoblasts. Med. Hypotheses 2007, 69, 629–636. [Google Scholar] [CrossRef]
- Ono, Y.; Sensui, H.; Sakamoto, Y.; Nagatomi, R. Knockdown of hypoxia-inducible factor-1alpha by siRNA inhibits C2C12 myoblast differentiation. J. Cell. Biochem. 2006, 98, 642–649. [Google Scholar] [CrossRef]
- Kubis, H.P.; Hanke, N.; Scheibe, R.J.; Gros, G. Accumulation and nuclear import of HIF1 alpha during high and low oxygen concentration in skeletal muscle cells in primary culture. Biochim. Biophys. Acta 2005, 1745, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Kim, D.; Rhee, J.; Seo, J.Y.; Park, I.; Kim, J.H.; Lee, D.; Lee, W.; Kim, Y.L.; Yoo, K.; et al. Baf155 regulates skeletal muscle metabolism via HIF-1a signaling. PLoS Biol. 2023, 21, e3002192. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Liu, Y.H.; Li, Y.Y.; Lu, Y.; Xu, H.Y. A Transcriptomic Analysis of Physiological Significance of Hypoxia-inducible Factor-1α in Myogenesis and Carbohydrate Metabolism of Genioglossus in Mice. Chin. Med. J. 2017, 130, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.; Simon, M.C. Hypoxia-inducible factors, stem cells, and cancer. Cell 2007, 129, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Kitakaze, T.; Sugihira, T.; Kameyama, H.; Maruchi, A.; Kobayashi, Y.; Harada, N.; Yamaji, R. Carotenoid transporter CD36 expression depends on hypoxia-inducible factor-1α in mouse soleus muscles. J. Clin. Biochem. Nutr. 2022, 71, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Settelmeier, S.; Schreiber, T.; Mäki, J.; Byts, N.; Koivunen, P.; Myllyharju, J.; Fandrey, J.; Winning, S. Prolyl hydroxylase domain 2 reduction enhances skeletal muscle tissue regeneration after soft tissue trauma in mice. PLoS ONE 2020, 15, e0233261. [Google Scholar] [CrossRef]
- Cicchillitti, L.; Di Stefano, V.; Isaia, E.; Crimaldi, L.; Fasanaro, P.; Ambrosino, V.; Antonini, A.; Capogrossi, M.C.; Gaetano, C.; Piaggio, G.; et al. Hypoxia-inducible factor 1-α induces miR-210 in normoxic differentiating myoblasts. J. Biol. Chem. 2012, 287, 44761–44771. [Google Scholar] [CrossRef]
- Wagatsuma, A.; Kotake, N.; Yamada, S. Spatial and temporal expression of hypoxia-inducible factor-1α during myogenesis in vivo and in vitro. Mol. Cell. Biochem. 2011, 347, 145–155. [Google Scholar] [CrossRef]
- Mounier, R.; Pedersen, B.K.; Plomgaard, P. Muscle-specific expression of hypoxia-inducible factor in human skeletal muscle. Exp. Physiol. 2010, 95, 899–907. [Google Scholar] [CrossRef]
- Stroka, D.M.; Burkhardt, T.; Desbaillets, I.; Wenger, R.H.; Neil, D.A.; Bauer, C.; Gassmann, M.; Candinas, D. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 2001, 15, 2445–2453. [Google Scholar] [CrossRef] [PubMed]
- Majmundar, A.J.; Lee, D.S.; Skuli, N.; Mesquita, R.C.; Kim, M.N.; Yodh, A.G.; Nguyen-McCarty, M.; Li, B.; Simon, M.C. HIF modulation of Wnt signaling regulates skeletal myogenesis in vivo. Development 2015, 142, 2405–2412. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, M.V.; Zheng, X.; Pereira, T.; Gradin, K.; Jin, S.; Lundkvist, J.; Ruas, J.L.; Poellinger, L.; Lendahl, U.; Bondesson, M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 2005, 9, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, H.; Cui, Y.; Li, W.; Zhang, S.; Chen, Y. Study on the Mechanism of Capillary Leakage Caused by Hypoxia-Inducible Factor-1α through Inducing High Expression of Matrix Metalloproteinase-9. J. Oncol. 2021, 2021, 9130650. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Zheng, Y.L. Hypoxia promotes invasion of retinoblastoma cells in vitro by upregulating HIF-1α/MMP9 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5361–5369. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Jang, Y.S.; Min, S.Y.; Song, J.Y. Overexpression of MMP-9 and HIF-1α in Breast Cancer Cells under Hypoxic Conditions. J. Breast Cancer 2011, 14, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Lu, K.V.; Petritsch, C.; Liu, P.; Ganss, R.; Passegué, E.; Song, H.; Vandenberg, S.; Johnson, R.S.; Werb, Z.; et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell 2008, 13, 206–220. [Google Scholar] [CrossRef]
- Sassoli, C.; Nosi, D.; Tani, A.; Chellini, F.; Mazzanti, B.; Quercioli, F.; Zecchi-Orlandini, S.; Formigli, L. Defining the role of mesenchymal stromal cells on the regulation of matrix metalloproteinases in skeletal muscle cells. Exp. Cell Res. 2014, 323, 297–313. [Google Scholar] [CrossRef]
- Nowak, E.; Gawor, M.; Ciemerych, M.A.; Zimowska, M. Silencing of gelatinase expression delays myoblast differentiation in vitro. Cell Biol. Int. 2018, 42, 373–382. [Google Scholar] [CrossRef]
- Rebalka, I.A.; Monaco, C.M.F.; Varah, N.E.; Berger, T.; D’souza, D.M.; Zhou, S.; Mak, T.W.; Hawke, T.J. Loss of the adipokine lipocalin-2 impairs satellite cell activation and skeletal muscle regeneration. Am. J. Physiol. Cell Physiol. 2018, 315, C714–C721. [Google Scholar] [CrossRef]
- Zimowska, M.; Swierczynska, M.; Ciemerych, M.A. Nuclear MMP-9 role in the regulation of rat skeletal myoblasts proliferation. Biol. Cell 2013, 105, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Squecco, R.; Chellini, F.; Idrizaj, E.; Tani, A.; Garella, R.; Pancani, S.; Pavan, P.; Bambi, F.; Zecchi-Orlandini, S.; Sassoli, C. Platelet-Rich Plasma Modulates Gap Junction Functionality and Connexin 43 and 26 Expression During TGF-β1-Induced Fibroblast to Myofibroblast Transition: Clues for Counteracting Fibrosis. Cells 2020, 9, 1199. [Google Scholar] [CrossRef] [PubMed]
- Sassoli, C.; Vallone, L.; Tani, A.; Chellini, F.; Nosi, D.; Zecchi-Orlandini, S. Combined use of bone marrow-derived mesenchymal stromal cells (BM-MSCs) and platelet rich plasma (PRP) stimulates proliferation and differentiation of myoblasts in vitro: New therapeutic perspectives for skeletal muscle repair/regeneration. Cell Tissue Res. 2018, 372, 549–570. [Google Scholar] [CrossRef] [PubMed]
- Bernacchioni, C.; Ghini, V.; Squecco, R.; Idrizaj, E.; Garella, R.; Puliti, E.; Cencetti, F.; Bruni, P.; Donati, C. Role of Sphingosine 1-Phosphate Signalling Axis in Muscle Atrophy Induced by TNFα in C2C12 Myotubes. Int. J. Mol. Sci. 2021, 22, 1280. [Google Scholar] [CrossRef]
- Formigli, L.; Sassoli, C.; Squecco, R.; Bini, F.; Martinesi, M.; Chellini, F.; Luciani, G.; Sbrana, F.; Zecchi-Orlandini, S.; Francini, F.; et al. Regulation of transient receptor potential canonical channel 1 (TRPC1) by sphingosine 1-phosphate in C2C12 myoblasts and its relevance for a role of mechanotransduction in skeletal muscle differentiation. J. Cell Sci. 2009, 122, 1322–1333. [Google Scholar] [CrossRef]
- Martella, D.; Mannelli, M.; Squecco, R.; Garella, R.; Idrizaj, E.; Antonioli, D.; Laus, M.; Wiersma, D.S.; Gamberi, T.; Paoli, P.; et al. Cell instructive Liquid Crystalline Networks for myotube formation. iScience 2021, 24, 103077. [Google Scholar] [CrossRef]
- Sassoli, C.; Pini, A.; Chellini, F.; Mazzanti, B.; Nistri, S.; Nosi, D.; Saccardi, R.; Quercioli, F.; Zecchi-Orlandini, S.; Formigli, L. Bone marrow mesenchymal stromal cells stimulate skeletal myoblast proliferation through the paracrine release of VEGF. PLoS ONE 2012, 7, e37512. [Google Scholar] [CrossRef]
- Luo, D.; Renault, V.M.; Rando, T.A. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin. Cell Dev. Biol. 2005, 16, 612–622. [Google Scholar] [CrossRef]
- Formigli, L.; Meacci, E.; Sassoli, C.; Squecco, R.; Nosi, D.; Chellini, F.; Naro, F.; Francini, F.; Zecchi-Orlandini, S. Cytoskeleton/stretch-activated ion channel interaction regulates myogenic differentiation of skeletal myoblasts. J. Cell. Physiol. 2007, 211, 296–306. [Google Scholar] [CrossRef]
- Sin, J.; Andres, A.M.; Taylor, D.J.; Weston, T.; Hiraumi, Y.; Stotland, A.; Kim, B.J.; Huang, C.; Doran, K.S.; Gottlieb, R.A. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 2016, 12, 369–380. [Google Scholar] [CrossRef]
- Wagatsuma, A.; Sakuma, K. Mitochondria as a potential regulator of myogenesis. Sci. World J. 2013, 2013, 593267. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology 2009, 24, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Summermatter, S.; Santos, G.; Pérez-Schindler, J.; Handschin, C. Skeletal muscle PGC-1α controls whole-body lactate homeostasis through estrogen-related receptor α-dependent activation of LDH B and repression of LDH A. Proc. Natl. Acad. Sci. USA 2013, 110, 8738–8743. [Google Scholar] [CrossRef] [PubMed]
- Hood, D.A.; Memme, J.M.; Oliveira, A.N.; Triolo, M. Maintenance of Skeletal Muscle Mitochondria in Health, Exercise, and Aging. Annu. Rev. Physiol. 2019, 81, 19–41. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.; Quan, L.; Lai, X.; Lang, L.; Li, F.; Yang, X.; Fu, Y.; Feng, S.; Yi, X.; Zhu, C.; et al. VEGFB Promotes Myoblasts Proliferation and Differentiation through VEGFR1-PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2021, 22, 13352. [Google Scholar] [CrossRef]
- Arsic, N.; Zacchigna, S.; Zentilin, L.; Ramirez-Correa, G.; Pattarini, L.; Salvi, A.; Sinagra, G.; Giacca, M. Vascular endothelial growth factor stimulates skeletal muscle regeneration in vivo. Mol. Ther. 2004, 10, 844–854. [Google Scholar] [CrossRef]
- Germani, A.; Di Carlo, A.; Mangoni, A.; Straino, S.; Giacinti, C.; Turrini, P.; Biglioli, P.; Capogrossi, M.C. Vascular endothelial growth factor modulates skeletal myoblast function. Am. J. Pathol. 2003, 163, 1417–1428. [Google Scholar] [CrossRef]
- Basic, V.T.; Jacobsen, A.; Sirsjö, A.; Abdel-Halim, S.M. TNF stimulation induces VHL overexpression and impairs angiogenic potential in skeletal muscle myocytes. Int. J. Mol. Med. 2014, 34, 228–236. [Google Scholar] [CrossRef]
- Rhoads, R.P.; Johnson, R.M.; Rathbone, C.R.; Liu, X.; Temm-Grove, C.; Sheehan, S.M.; Hoying, J.B.; Allen, R.E. Satellite cell-mediated angiogenesis in vitro coincides with a functional hypoxia-inducible factor pathway. Am. J. Physiol. Cell Physiol. 2009, 296, C1321–C1328. [Google Scholar] [CrossRef]
- Hirota, K.; Semenza, G.L. Regulation of angiogenesis by hypoxia-inducible factor 1. Crit. Rev. Oncol. Hematol. 2006, 59, 15–26. [Google Scholar] [CrossRef]
- Olson, N.; van der Vliet, A. Interactions between nitric oxide and hypoxia-inducible factor signaling pathways in inflammatory disease. Nitric Oxide 2011, 25, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Déry, M.A.; Michaud, M.D.; Richard, D.E. Hypoxia-inducible factor 1: Regulation by hypoxic and non-hypoxic activators. Int. J. Biochem. Cell Biol. 2005, 37, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Sibisi, N.C.; Snyman, C.; Myburgh, K.H.; Niesler, C.U. Evaluating the role of nitric oxide in myogenesis in vitro. Biochimie 2022, 196, 216–224. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, K.A.; Cocchiglia, S.; Zhdanov, A.V.; Tambuwala, M.M.; Cummins, E.P.; Monfared, M.; Agbor, T.A.; Garvey, J.F.; Papkovsky, D.B.; Taylor, C.T.; et al. PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2188–2193. [Google Scholar] [CrossRef] [PubMed]
- Villa, J.C.; Chiu, D.; Brandes, A.H.; Escorcia, F.E.; Villa, C.H.; Maguire, W.F.; Hu, C.J.; de Stanchina, E.; Simon, M.C.; Sisodia, S.S.; et al. Nontranscriptional role of Hif-1 α in activation of γ-secretase and notch signaling in breast cancer. Cell Rep. 2014, 8, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Sakagami, H.; Makino, Y.; Mizumoto, K.; Isoe, T.; Takeda, Y.; Watanabe, J.; Fujita, Y.; Takiyama, Y.; Abiko, A.; Haneda, M. Loss of HIF-1α impairs GLUT4 translocation and glucose uptake by the skeletal muscle cells. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1065–E1076. [Google Scholar] [CrossRef] [PubMed]
- Hubbi, M.E.; Kshitiz, G.D.M.; Rey, S.; Wong, C.C.; Luo, W.; Kim, D.H.; Dang, C.V.; Levchenko, A.; Semenza, G.L. A nontranscriptional role for HIF-1α as a direct inhibitor of DNA replication. Sci. Signal. 2013, 6, ra10. [Google Scholar] [CrossRef]
- Schmidt, M.; Schüler, S.C.; Hüttner, S.S.; von Eyss, B.; von Maltzahn, J. Adult stem cells at work: Regenerating skeletal muscle. Cell. Mol. Life Sci. 2019, 76, 2559–2570. [Google Scholar] [CrossRef]
- Yun, Z.; Lin, Q.; Giaccia, A.J. Adaptive myogenesis under hypoxia. Mol. Cell. Biol. 2005, 25, 3040–3055. [Google Scholar] [CrossRef]
- Zimowska, M.; Olszynski, K.H.; Swierczynska, M.; Streminska, W.; Ciemerych, M.A. Decrease of MMP-9 activity improves soleus muscle regeneration. Tissue Eng. Part A 2012, 18, 1183–1192. [Google Scholar] [CrossRef]
- Kann, A.P.; Hung, M.; Wang, W.; Nguyen, J.; Gilbert, P.M.; Wu, Z.; Krauss, R.S. An injury-responsive Rac-to-Rho GTPase switch drives activation of muscle stem cells through rapid cytoskeletal remodeling. Cell Stem Cell 2022, 29, 933–947.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Z.G.; Taiyab, A.; West-Mays, J.A. MMP9 Differentially Regulates Proteins Involved in Actin Polymerization and Cell Migration during TGF-β-Induced EMT in the Lens. Int. J. Mol. Sci. 2021, 22, 11988. [Google Scholar] [CrossRef] [PubMed]
- Bernacchioni, C.; Squecco, R.; Gamberi, T.; Ghini, V.; Schumacher, F.; Mannelli, M.; Garella, R.; Idrizaj, E.; Cencetti, F.; Puliti, E.; et al. S1P Signalling Axis Is Necessary for Adiponectin-Directed Regulation of Electrophysiological Properties and Oxidative Metabolism in C2C12 Myotubes. Cells 2022, 11, 713. [Google Scholar] [CrossRef] [PubMed]
- Muratore, M.; Srsen, V.; Waterfall, M.; Downes, A.; Pethig, R. Biomarker-free dielectrophoretic sorting of differentiating myoblast multipotent progenitor cells and their membrane analysis by Raman spectroscopy. Biomicrofluidics 2012, 6, 34113. [Google Scholar] [CrossRef] [PubMed]
- Meacci, E.; Bini, F.; Sassoli, C.; Martinesi, M.; Squecco, R.; Chellini, F.; Zecchi-Orlandini, S.; Francini, F.; Formigli, L. Functional interaction between TRPC1 channel and connexin-43 protein: A novel pathway underlying S1P action on skeletal myogenesis. Cell. Mol. Life Sci. 2010, 67, 4269–4285. [Google Scholar] [CrossRef]
- Leikina, E.; Gamage, D.G.; Prasad, V.; Goykhberg, J.; Crowe, M.; Diao, J.; Kozlov, M.M.; Chernomordik, L.V.; Millay, D.P. Myomaker and Myomerger Work Independently to Control Distinct Steps of Membrane Remodeling during Myoblast Fusion. Dev. Cell 2018, 46, 767–780.e7. [Google Scholar] [CrossRef]
- Muller, M.; Schick, M. An alternate path for fusion and its exploration by field-theoretic means. Curr. Top. Membr. 2011, 68, 295–323. [Google Scholar] [CrossRef]
- Chernomordik, L.V.; Kozlov, M.M. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003, 72, 175–207. [Google Scholar] [CrossRef]
- Shimahara, T.; Bournaud, R. Barium currents in developing skeletal muscle cells of normal and mutant mice foetuses with ‘muscular dysgenesis’. Cell Calcium 1991, 12, 727–733. [Google Scholar] [CrossRef]
- Tamayo, T.; Grajales, L.; García, J. Commitment of satellite cells expressing the calcium channel α2δ1 subunit to the muscle lineage. J. Signal Transduct. 2012, 2012, 460842. [Google Scholar] [CrossRef]
- Grajales, L.; Lach, L.E.; Janisch, P.; Geenen, D.L.; García, J. Temporal Expression of Calcium Channel Subunits in Satellite Cells and Bone Marrow Mesenchymal Cells. Stem Cell Rev. Rep. 2015, 11, 408–422. [Google Scholar] [CrossRef] [PubMed]
- García, K.; Nabhani, T.; García, J. The calcium channel α2/δ1 subunit is involved in extracellular signaling. J. Physiol. 2008, 586, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Weigand, L.; Lu, W.; Sylvester, J.T.; Semenza, G.L.; Shimoda, L.A. Hypoxia Inducible Factor 1 Mediates Hypoxia-Induced TRPC Expression and Elevated Intracellular Ca2+ in Pulmonary Arterial Smooth Muscle Cells. Circ. Res. 2006, 98, 1528–1537. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Mimura, I.; Kurata, Y.; Tanaka, T.; Nangaku, M. Dznep, a histone modification inhibitor, inhibits HIF1α binding to TIMP2 gene and suppresses TIMP2 expression under hypoxia. Physiol. Rep. 2023, 11, e15810. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Gu, C.; Shen, Q.; Liu, J.; Zhou, Y.; Jin, Z.; Xiong, W.; Zhou, Y.; Tan, W. Hypoxia alleviates dexamethasone-induced inhibition of angiogenesis in cocultures of HUVECs and rBMSCs via HIF-1α. Stem Cell Res. Ther. 2020, 11, 343. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, W.; Zhang, F.; Li, P.L.; Boini, K.M.; Yi, F.; Li, N. Hypoxia inducible factor-1α mediates the profibrotic effect of albumin in renal tubular cells. Sci. Rep. 2017, 7, 15878. [Google Scholar] [CrossRef]
- Takahara, Y.; Tokunou, T.; Kojima, H.; Hirooka, Y.; Ichiki, T. Deletion of hypoxia-inducible factor-1α in myeloid lineage exaggerates angiotensin II-induced formation of abdominal aortic aneurysm. Clin. Sci. 2017, 131, 609–620. [Google Scholar] [CrossRef]
- Kai, A.K.; Chan, L.K.; Lo, R.C.; Lee, J.M.; Wong, C.C.; Wong, J.C.; Ng, I.O. Down-regulation of TIMP2 by HIF-1α/miR-210/HIF-3α regulatory feedback circuit enhances cancer metastasis in hepatocellular carcinoma. Hepatology 2016, 64, 473–487. [Google Scholar] [CrossRef]
- Schelter, F.; Halbgewachs, B.; Bäumler, P.; Neu, C.; Görlach, A.; Schrötzlmair, F.; Krüger, A. Tissue inhibitor of metalloproteinases-1-induced scattered liver metastasis is mediated by hypoxia-inducible factor-1α. Clin. Exp. Metastasis 2011, 28, 91–99. [Google Scholar] [CrossRef]
- Leeman, M.F.; Curran, S.; Murray, G.I. The structure, regulation, and function of human matrix metalloproteinase-13. Crit. Rev. Biochem. Mol. Biol. 2002, 37, 149–166. [Google Scholar] [CrossRef]
- Lei, H.; Leong, D.; Smith, L.R.; Barton, E.R. Matrix metalloproteinase 13 is a new contributor to skeletal muscle regeneration and critical for myoblast migration. Am. J. Physiol. Cell Physiol. 2013, 305, C529–C538. [Google Scholar] [CrossRef] [PubMed]










| Primary Antibody Name | Company | Code | Dilution |
|---|---|---|---|
| anti-Ki67 rabbit polyclonal | Abcam, Cambridge, UK | ab15580 | 1:100 (IF) |
| anti-Notch-1 rabbit monoclonal | Abcam | ab52627 | 1:100 (IF) 1:1000 (WB) |
| anti-MyoD (M-318) rabbit polyclonal | Santa Cruz Biotechnology | sc-760 | 1:50 (IF) 1:500 (WB) |
| anti-myogenin (F5D) mouse monoclonal | Santa Cruz Biotechnology | sc-12732 | 1:50 (IF) |
| anti-α-sarcomeric actin mouse monoclonal | DakoCytomation, Carpinteria, CA, USA | M 0874 | 1:100 (IF) |
| anti- peroxisome proliferator-activated receptor-gamma coactivator (PGC)-1α mouse monoclonal | Santa Cruz Biotechnology | sc-518025 | 1:100 (IF) |
| anti-HIF-1α rabbit polyclonal | Santa Cruz Biotechnology | sc-10790 | 1:100 (IF) 1:1000 (WB) |
| anti-MMP-9 rabbit polyclonal | Bioss Antibodies, Woburn, MA, USA | bs-0397R | 1:100 (IF) 1:1000 (WB) |
| anti-Pax7 mouse monoclonal | Santa Cruz Biotechnology | sc-81648 | 1:100 (IF) |
| anti-succinate dehydrogenase complex iron sulfur subunit-B (SDH-B) rabbit polyclonal | Sigma-Aldrich | SAB2102103 | 1:1000 (WB) |
| anti-lactate dehydrogenase (LDH)-A (E-9) mouse monoclonal | Santa Cruz Biotechnology | sc-137243 | 1:1000 (WB) |
| anti-α-tubulin rabbit polyclonal | GeneTex, Prodotti Gianni, Milano, Italy | GTX112141 | 1:1000 (WB) |
| Condition | Cm (pF) | Gm/Cm (nS/pF) |
|---|---|---|
| WT PM | 23.56 ± 4.20 n = 5 | 1.29 × 10−2 ± 0.77 × 10−2 n = 10 |
| SCR-siRNA PM (T0) | 15.51 ± 4.43 n = 3 | 2.18 × 10−2 ± 1.79 × 10−2 n = 4 |
| HIF-1α-siRNA PM (T0) | 19.65 ± 6.68 n = 4 | 2.82 × 10−2 ± 1.97 × 10−2 n = 4 |
| SB-3CT PM 24 h | 25.08 ± 17.06 n = 6 | 3.78 × 10−2 ± 3.10 × 10−2 n = 5 |
| WT DM 24 h | 41.87 ± 19.62 * n = 10 | 3.04 × 10−2 ± 2.21 × 10−2 * n = 10 |
| SCR-siRNA DM 24 h | 40.58 ± 25.22 * n = 7 | 4.11 × 10−2 ± 2,09 × 10−2 * n = 6 |
| HIF-1α-siRNA DM 24 h | 20.08 ± 7.02 # n = 5 | 1.27 × 10−2 ± 1.03 × 10−2 # n = 11 |
| SB-3CT DM 24 h | 19.68 ± 10.81 # n = 5 | 0.93 × 10−2 ± 0.82 × 10−2 # n = 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chellini, F.; Tani, A.; Parigi, M.; Palmieri, F.; Garella, R.; Zecchi-Orlandini, S.; Squecco, R.; Sassoli, C. HIF-1α/MMP-9 Axis Is Required in the Early Phases of Skeletal Myoblast Differentiation under Normoxia Condition In Vitro. Cells 2023, 12, 2851. https://doi.org/10.3390/cells12242851
Chellini F, Tani A, Parigi M, Palmieri F, Garella R, Zecchi-Orlandini S, Squecco R, Sassoli C. HIF-1α/MMP-9 Axis Is Required in the Early Phases of Skeletal Myoblast Differentiation under Normoxia Condition In Vitro. Cells. 2023; 12(24):2851. https://doi.org/10.3390/cells12242851
Chicago/Turabian StyleChellini, Flaminia, Alessia Tani, Martina Parigi, Francesco Palmieri, Rachele Garella, Sandra Zecchi-Orlandini, Roberta Squecco, and Chiara Sassoli. 2023. "HIF-1α/MMP-9 Axis Is Required in the Early Phases of Skeletal Myoblast Differentiation under Normoxia Condition In Vitro" Cells 12, no. 24: 2851. https://doi.org/10.3390/cells12242851