Short Linear Motifs in Colorectal Cancer Interactome and Tumorigenesis
Abstract
:1. Introduction
2. Short Linear Motifs and Their Emerging Role in Cell Biology and Cancer
3. In Silico Methods to Study PPI Networks
4. In Silico Approaches and Tools to Characterize SLiMs in PPI Networks
5. SLiMs in CRC Molecular Networks
5.1. SLiMs in CRC Signaling Pathways and Tumorigenesis
5.2. SLiMs in CRC Hallmarks: A Case Study
5.3. SLiMs in CRC-Related Microbiome
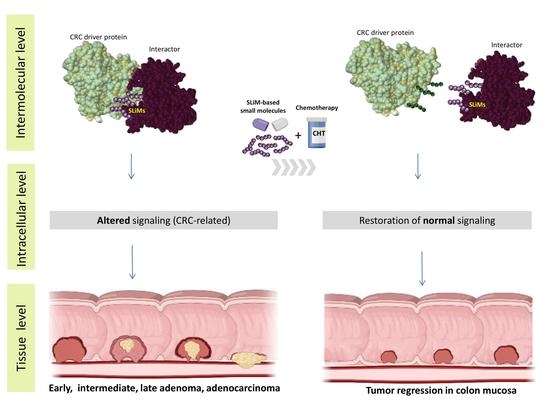
6. Potential Small-Molecule Anticancer Drugs Based on SLiMs and Short Peptides in CRC: Where Do We Stand?
6.1. Pharmacological Suitability of SLiMs and Short Peptides as Anticancer Drugs
6.2. Short Peptides as Potential Anti-CRC Drugs
6.3. Short Peptides in Clinical Studies
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hossain, M.S.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, D.J.; John, A.; Lim, Y.C.; Kibria, K.M.K.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef]
- Jasperson, K.W.; Tuohy, T.M.; Neklason, D.W.; Burt, R.W. Hereditary and Familial Colon Cancer. Gastroenterology 2010, 138, 2044–2058. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global Burden of Colorectal Cancer: Emerging Trends, Risk Factors and Prevention Strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Olkinuora, A.P.; Peltomäki, P.T.; Aaltonen, L.A.; Rajamäki, K. From APC to the Genetics of Hereditary and Familial Colon Cancer Syndromes. Hum. Mol. Genet. 2021, 30, R206–R224. [Google Scholar] [CrossRef]
- Bhandari, A.; Woodhouse, M.; Gupta, S. Colorectal Cancer Is a Leading Cause of Cancer Incidence and Mortality among Adults Younger than 50 Years in the USA: A SEER-Based Analysis with Comparison to Other Young-Onset Cancers. J. Investig. Med. 2017, 65, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yang, M. Molecular Network of Colorectal Cancer and Current Therapeutic Options. Front. Oncol. 2022, 12, 852927. [Google Scholar] [CrossRef]
- Perkins, J.R.; Diboun, I.; Dessailly, B.H.; Lees, J.G.; Orengo, C. Transient Protein-Protein Interactions: Structural, Functional, and Network Properties. Structure 2010, 18, 1233–1243. [Google Scholar] [CrossRef]
- Davey, N.E.; Van Roey, K.; Weatheritt, R.J.; Toedt, G.; Uyar, B.; Altenberg, B.; Budd, A.; Diella, F.; Dinkel, H.; Gibson, T.J. Attributes of Short Linear Motifs. Mol. Biosyst. 2012, 8, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, B.; Simon, I.; Dosztányi, Z. Prediction of Protein Binding Regions in Disordered Proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef]
- Neduva, V.; Russell, R.B. Peptides Mediating Interaction Networks: New Leads at Last. Curr. Opin. Biotechnol. 2006, 17, 465–471. [Google Scholar] [CrossRef]
- Diella, F.; Haslam, N.; Chica, C.; Budd, A.; Michael, S.; Brown, N.P.; Trave, G.; Gibson, T.J. Understanding Eukaryotic Linear Motifs and Their Role in Cell Signaling and Regulation. Front. Biosci. 2008, 13, 6580–6603. [Google Scholar] [CrossRef]
- Davey, N.E.; Edwards, R.J.; Shields, D.C. Computational Identification and Analysis of Protein Short Linear Motifs. Front. Biosci. (Landmark Ed) 2010, 15, 801–825. [Google Scholar] [CrossRef]
- Marongiu, L.; Allgayer, H. Viruses in Colorectal Cancer. Mol. Oncol. 2022, 16, 1423–1450. [Google Scholar] [CrossRef] [PubMed]
- Sai Krishna, A.V.S.; Sinha, S.; Donakonda, S. Virus-Host Interaction Analysis in Colorectal Cancer Identifies Core Virus Network Signature and Small Molecules. Comput. Struct. Biotechnol. J. 2022, 20, 4025–4039. [Google Scholar] [CrossRef]
- Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Mészáros, B.; Sámano-Sánchez, H.; Zeke, A.; Dobson, L.; Lazar, T.; Örd, M.; Nagpal, A.; et al. The Eukaryotic Linear Motif Resource: 2022 Release. Nucleic Acids Res. 2022, 50, D497–D508. [Google Scholar] [CrossRef] [PubMed]
- Aouacheria, A.; Combet, C.; Tompa, P.; Hardwick, J.M. Redefining the BH3 Death Domain as a “Short Linear Motif”. Trends Biochem. Sci. 2015, 40, 736–748. [Google Scholar] [CrossRef]
- Petsalaki, E.; Russell, R.B. Peptide-Mediated Interactions in Biological Systems: New Discoveries and Applications. Curr. Opin. Biotechnol. 2008, 19, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Clerc, I.; Sagar, A.; Barducci, A.; Sibille, N.; Bernadó, P.; Cortés, J. The Diversity of Molecular Interactions Involving Intrinsically Disordered Proteins: A Molecular Modeling Perspective. Comput. Struct. Biotechnol. J. 2021, 19, 3817–3828. [Google Scholar] [CrossRef]
- Kim, I.; Lee, H.; Han, S.K.; Kim, S. Linear Motif-Mediated Interactions Have Contributed to the Evolution of Modularity in Complex Protein Interaction Networks. PLoS Comput. Biol. 2014, 10, e1003881. [Google Scholar] [CrossRef]
- Verschueren, E.; Vanhee, P.; van der Sloot, A.M.; Serrano, L.; Rousseau, F.; Schymkowitz, J. Protein Design with Fragment Databases. Curr. Opin. Struct. Biol. 2011, 21, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Sologova, S.S.; Zavadskiy, S.P.; Mokhosoev, I.M.; Moldogazieva, N.T. Short Linear Motifs Orchestrate Functioning of Human Proteins during Embryonic Development, Redox Regulation, and Cancer. Metabolites 2022, 12, 464. [Google Scholar] [CrossRef] [PubMed]
- Hartooni, N.; Sung, J.; Jain, A.; Morgan, D.O. Single-Molecule Analysis of Specificity and Multivalency in Binding of Short Linear Substrate Motifs to the APC/C. Nat. Commun. 2022, 13, 341. [Google Scholar] [CrossRef]
- Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Davey, N.E.; Gibson, T.J. Proteome-Wide Analysis of Human Disease Mutations in Short Linear Motifs: Neglected Players in Cancer? Mol. Biosyst. 2014, 10, 2626–2642. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. Unstructural Biology Coming of Age. Curr. Opin. Struct. Biol. 2011, 21, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chini, C.C.S.; He, M.; Mer, G.; Chen, J. The BRCT Domain Is a Phospho-Protein Binding Domain. Science 2003, 302, 639–642. [Google Scholar] [CrossRef] [PubMed]
- Clapperton, J.A.; Manke, I.A.; Lowery, D.M.; Ho, T.; Haire, L.F.; Yaffe, M.B.; Smerdon, S.J. Structure and Mechanism of BRCA1 BRCT Domain Recognition of Phosphorylated BACH1 with Implications for Cancer. Nat. Struct. Mol. Biol. 2004, 11, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Al-Sukhni, W.; Rothenmund, H.; Borgida, A.E.; Zogopoulos, G.; O’Shea, A.-M.; Pollett, A.; Gallinger, S. Germline BRCA1 Mutations Predispose to Pancreatic Adenocarcinoma. Hum. Genet. 2008, 124, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Jubb, H.; Blundell, T.L. Phosphopeptide Interactions with BRCA1 BRCT Domains: More than Just a Motif. Prog. Biophys. Mol. Biol. 2015, 117, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Cantor, S.; Drapkin, R.; Zhang, F.; Lin, Y.; Han, J.; Pamidi, S.; Livingston, D.M. The BRCA1-Associated Protein BACH1 Is a DNA Helicase Targeted by Clinically Relevant Inactivating Mutations. Proc. Natl. Acad. Sci. USA 2004, 101, 2357–2362. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Terent’ev, A.A.; Shaĭtan, K.V. Relationship between structure and function of alpha-fetoprotein: Conformational status and biological activity. Biomed. Khim. 2005, 51, 127–151. [Google Scholar]
- Zhu, Z.; West, G.R.; Wang, D.C.; Collins, A.B.; Xiao, H.; Bai, Q.; Mesfin, F.B.; Wakefield, M.R.; Fang, Y. AFP Peptide (AFPep) as a Potential Growth Factor for Prostate Cancer. Med. Oncol. 2021, 39, 2. [Google Scholar] [CrossRef] [PubMed]
- Moldogazieva, N.T.; Zavadskiy, S.P.; Sologova, S.S.; Mokhosoev, I.M.; Terentiev, A.A. Predictive Biomarkers for Systemic Therapy of Hepatocellular Carcinoma. Expert Rev. Mol. Diagn. 2021, 21, 1147–1164. [Google Scholar] [CrossRef] [PubMed]
- Moldogazieva, N.T.; Shaitan, K.V.; Antonov, M.Y.; Mokhosoev, I.M.; Levtsova, O.V.; Terentiev, A.A. Human EGF-Derived Direct and Reverse Short Linear Motifs: Conformational Dynamics Insight into the Receptor-Binding Residues. J. Biomol. Struct. Dyn. 2018, 36, 1286–1305. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Borcherds, W.; Wu, S.; Becker, A.; Schonbrunn, E.; Daughdrill, G.W.; Chen, J. Autoinhibition of MDMX by Intramolecular P53 Mimicry. Proc. Natl. Acad. Sci. USA 2015, 112, 4624–4629. [Google Scholar] [CrossRef] [PubMed]
- Fenton, M.; Borcherds, W.; Chen, L.; Anbanandam, A.; Levy, R.; Chen, J.; Daughdrill, G. The MDMX Acidic Domain Uses Allovalency to Bind Both P53 and MDMX. J. Mol. Biol. 2022, 434, 167844. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.J.; Davey, N.E.; O’Brien, K.; Shields, D.C. Interactome-Wide Prediction of Short, Disordered Protein Interaction Motifs in Humans. Mol. Biosyst. 2012, 8, 282–295. [Google Scholar] [CrossRef] [PubMed]
- Halakou, F.; Gursoy, A.; Keskin, O. Embedding Alternative Conformations of Proteins in Protein-Protein Interaction Networks. Methods Mol. Biol. 2020, 2074, 113–124. [Google Scholar] [CrossRef]
- Ivanov, A.A.; Revennaugh, B.; Rusnak, L.; Gonzalez-Pecchi, V.; Mo, X.; Johns, M.A.; Du, Y.; Cooper, L.A.D.; Moreno, C.S.; Khuri, F.R.; et al. The OncoPPi Portal: An Integrative Resource to Explore and Prioritize Protein-Protein Interactions for Cancer Target Discovery. Bioinformatics 2018, 34, 1183–1191. [Google Scholar] [CrossRef]
- Ivanov, A.A. Explore Protein–Protein Interactions for Cancer Target Discovery Using the OncoPPi Portal. In Protein-Protein Interaction Networks: Methods and Protocols; Canzar, S., Ringeling, F.R., Eds.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2020; pp. 145–164. ISBN 978-1-4939-9873-9. [Google Scholar]
- Du, Y.; Cai, M.; Xing, X.; Ji, J.; Yang, E.; Wu, J. PINA 3.0: Mining Cancer Interactome. Nucleic Acids Res. 2021, 49, D1351–D1357. [Google Scholar] [CrossRef]
- Wu, J.; Vallenius, T.; Ovaska, K.; Westermarck, J.; Mäkelä, T.P.; Hautaniemi, S. Integrated Network Analysis Platform for Protein-Protein Interactions. Nat. Methods 2009, 6, 75–77. [Google Scholar] [CrossRef]
- Li, Y.; Ilie, L. SPRINT: Ultrafast Protein-Protein Interaction Prediction of the Entire Human Interactome. BMC Bioinform. 2017, 18, 485. [Google Scholar] [CrossRef]
- Philipp, O.; Osiewacz, H.D.; Koch, I. Path2PPI: An R Package to Predict Protein-Protein Interaction Networks for a Set of Proteins. Bioinformatics 2016, 32, 1427–1429. [Google Scholar] [CrossRef]
- Gueudré, T.; Baldassi, C.; Pagnani, A.; Weigt, M. Predicting Interacting Protein Pairs by Coevolutionary Paralog Matching. Methods Mol. Biol. 2020, 2074, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Zeng, H.; Wang, S.; Xu, J. A Web-Based Protocol for Interprotein Contact Prediction by Deep Learning. Methods Mol. Biol. 2020, 2074, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Furmanová, K.; Byška, J.; Gröller, E.M.; Viola, I.; Paleček, J.J.; Kozlíková, B. COZOID: Contact Zone Identifier for Visual Analysis of Protein-Protein Interactions. BMC Bioinform. 2018, 19, 125. [Google Scholar] [CrossRef]
- Peterson, L.X.; Togawa, Y.; Esquivel-Rodriguez, J.; Terashi, G.; Christoffer, C.; Roy, A.; Shin, W.-H.; Kihara, D. Modeling the Assembly Order of Multimeric Heteroprotein Complexes. PLoS Comput. Biol. 2018, 14, e1005937. [Google Scholar] [CrossRef]
- Loney, F.; Wu, G. Automation of ReactomeFIViz via CyREST API. F1000Research 2018, 7, 531. [Google Scholar] [CrossRef]
- Alcaraz, N.; Pauling, J.; Batra, R.; Barbosa, E.; Junge, A.; Christensen, A.G.L.; Azevedo, V.; Ditzel, H.J.; Baumbach, J. KeyPathwayMiner 4.0: Condition-Specific Pathway Analysis by Combining Multiple Omics Studies and Networks with Cytoscape. BMC Syst. Biol. 2014, 8, 99. [Google Scholar] [CrossRef]
- Biedermann, S.; Henzinger, M.; Schulz, C.; Schuster, B. Vienna Graph Clustering. In Protein-Protein Interaction Networks: Methods and Protocols; Canzar, S., Ringeling, F.R., Eds.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2020; pp. 215–231. ISBN 978-1-4939-9873-9. [Google Scholar]
- Re, A.; Lecca, P. On TD-WGcluster: Theoretical Foundations and Guidelines for the User. In Protein-Protein Interaction Networks: Methods and Protocols; Canzar, S., Ringeling, F.R., Eds.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2020; pp. 233–262. ISBN 978-1-4939-9873-9. [Google Scholar]
- Mamano, N.; Hayes, W.B. SANA: Simulated Annealing Far Outperforms Many Other Search Algorithms for Biological Network Alignment. Bioinformatics 2017, 33, 2156–2164. [Google Scholar] [CrossRef]
- Zhou, A.; Zhang, F.; Chen, J.Y. PEPPI: A Peptidomic Database of Human Protein Isoforms for Proteomics Experiments. BMC Bioinform. 2010, 11 (Suppl. 6), S7. [Google Scholar] [CrossRef]
- Kamburov, A.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB--a Database for Integrating Human Functional Interaction Networks. Nucleic Acids Res. 2009, 37, D623–D628. [Google Scholar] [CrossRef]
- Kotlyar, M.; Pastrello, C.; Sheahan, N.; Jurisica, I. Integrated Interactions Database: Tissue-Specific View of the Human and Model Organism Interactomes. Nucleic Acids Res. 2016, 44, D536–D541. [Google Scholar] [CrossRef] [PubMed]
- Alanis-Lobato, G.; Andrade-Navarro, M.A.; Schaefer, M.H. HIPPIE v2.0: Enhancing Meaningfulness and Reliability of Protein–Protein Interaction Networks. Nucleic Acids Res. 2017, 45, D408–D414. [Google Scholar] [CrossRef] [PubMed]
- Pagel, P.; Kovac, S.; Oesterheld, M.; Brauner, B.; Dunger-Kaltenbach, I.; Frishman, G.; Montrone, C.; Mark, P.; Stümpflen, V.; Mewes, H.-W.; et al. The MIPS Mammalian Protein-Protein Interaction Database. Bioinformatics 2005, 21, 832–834. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.-J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID Database: A Comprehensive Biomedical Resource of Curated Protein, Genetic, and Chemical Interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct Project--IntAct as a Common Curation Platform for 11 Molecular Interaction Databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef]
- Reys, V.; Labesse, G. SLiMAn: An Integrative Web Server for Exploring Short Linear Motif-Mediated Interactions in Interactomes. J. Proteome Res. 2022, 21, 1654–1663. [Google Scholar] [CrossRef]
- Groner, B.; Weber, A.; Mack, L. Increasing the Range of Drug Targets: Interacting Peptides Provide Leads for the Development of Oncoprotein Inhibitors. Bioengineered 2012, 3, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Amos-Binks, A.; Patulea, C.; Pitre, S.; Schoenrock, A.; Gui, Y.; Green, J.R.; Golshani, A.; Dehne, F. Binding Site Prediction for Protein-Protein Interactions and Novel Motif Discovery Using Re-Occurring Polypeptide Sequences. BMC Bioinform. 2011, 12, 225. [Google Scholar] [CrossRef]
- Mi, T.; Merlin, J.C.; Deverasetty, S.; Gryk, M.R.; Bill, T.J.; Brooks, A.W.; Lee, L.Y.; Rathnayake, V.; Ross, C.A.; Sargeant, D.P.; et al. Minimotif Miner 3.0: Database Expansion and Significantly Improved Reduction of False-Positive Predictions from Consensus Sequences. Nucleic Acids Res. 2012, 40, D252–D260. [Google Scholar] [CrossRef]
- Sarkar, D.; Jana, T.; Saha, S. LMPID: A Manually Curated Database of Linear Motifs Mediating Protein-Protein Interactions. Database (Oxford) 2015, 2015, bav014. [Google Scholar] [CrossRef]
- Encinar, J.A.; Fernandez-Ballester, G.; Sánchez, I.E.; Hurtado-Gomez, E.; Stricher, F.; Beltrao, P.; Serrano, L. ADAN: A Database for Prediction of Protein-Protein Interaction of Modular Domains Mediated by Linear Motifs. Bioinformatics 2009, 25, 2418–2424. [Google Scholar] [CrossRef]
- Dinkel, H.; Sticht, H. A Computational Strategy for the Prediction of Functional Linear Peptide Motifs in Proteins. Bioinformatics 2007, 23, 3297–3303. [Google Scholar] [CrossRef]
- Davey, N.E.; Haslam, N.J.; Shields, D.C.; Edwards, R.J. SLiMSearch 2.0: Biological Context for Short Linear Motifs in Proteins. Nucleic Acids Res. 2011, 39, W56–W60. [Google Scholar] [CrossRef]
- Dosztányi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web Server for the Prediction of Intrinsically Unstructured Regions of Proteins Based on Estimated Energy Content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Via, A.; Gould, C.M.; Gemünd, C.; Gibson, T.J.; Helmer-Citterich, M. A Structure Filter for the Eukaryotic Linear Motif Resource. BMC Bioinform. 2009, 10, 351. [Google Scholar] [CrossRef]
- Petsalaki, E.; Stark, A.; García-Urdiales, E.; Russell, R.B. Accurate Prediction of Peptide Binding Sites on Protein Surfaces. PLoS Comput. Biol. 2009, 5, e1000335. [Google Scholar] [CrossRef]
- Ramu, C. SIRW: A Web Server for the Simple Indexing and Retrieval System That Combines Sequence Motif Searches with Keyword Searches. Nucleic Acids Res. 2003, 31, 3771–3774. [Google Scholar] [CrossRef]
- Wang, G.; Matsuura, I.; He, D.; Liu, F. Transforming Growth Factor-{beta}-Inducible Phosphorylation of Smad3. J. Biol. Chem. 2009, 284, 9663–9673. [Google Scholar] [CrossRef]
- Koveitypour, Z.; Panahi, F.; Vakilian, M.; Peymani, M.; Seyed Forootan, F.; Nasr Esfahani, M.H.; Ghaedi, K. Signaling Pathways Involved in Colorectal Cancer Progression. Cell Biosci. 2019, 9, 97. [Google Scholar] [CrossRef]
- Ramesh, P.; Medema, J.P. BCL-2 Family Deregulation in Colorectal Cancer: Potential for BH3 Mimetics in Therapy. Apoptosis 2020, 25, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Bienz, M.; Clevers, H. Linking Colorectal Cancer to Wnt Signaling. Cell 2000, 103, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Dannappel, M.; Wan, C.; Firestein, R. Transcriptional Regulation of Wnt/β-Catenin Pathway in Colorectal Cancer. Cells 2020, 9, 2125. [Google Scholar] [CrossRef]
- Wnt Signaling in Colorectal Cancer: Pathogenic Role and Therapeutic Target-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/35836256/ (accessed on 3 August 2022).
- Singh, V.K.; Pacheco, I.; Uversky, V.N.; Smith, S.P.; MacLeod, R.J.; Jia, Z. Intrinsically Disordered Human C/EBP Homologous Protein Regulates Biological Activity of Colon Cancer Cells during Calcium Stress. J. Mol. Biol. 2008, 380, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Xu, G.; Schulman, B.A.; Jeffrey, P.D.; Harper, J.W.; Pavletich, N.P. Structure of a Beta-TrCP1-Skp1-Beta-Catenin Complex: Destruction Motif Binding and Lysine Specificity of the SCF(Beta-TrCP1) Ubiquitin Ligase. Mol. Cell 2003, 11, 1445–1456. [Google Scholar] [CrossRef]
- Lustig, B.; Behrens, J. The Wnt Signaling Pathway and Its Role in Tumor Development. J. Cancer Res. Clin. Oncol. 2003, 129, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Sadot, E.; Conacci-Sorrell, M.; Zhurinsky, J.; Shnizer, D.; Lando, Z.; Zharhary, D.; Kam, Z.; Ben-Ze’ev, A.; Geiger, B. Regulation of S33/S37 Phosphorylated Beta-Catenin in Normal and Transformed Cells. J. Cell Sci. 2002, 115, 2771–2780. [Google Scholar] [CrossRef] [PubMed]
- Provost, E.; McCabe, A.; Stern, J.; Lizardi, I.; D’Aquila, T.G.; Rimm, D.L. Functional Correlates of Mutation of the Asp32 and Gly34 Residues of Beta-Catenin. Oncogene 2005, 24, 2667–2676. [Google Scholar] [CrossRef]
- Welcker, M.; Clurman, B.E. FBW7 Ubiquitin Ligase: A Tumour Suppressor at the Crossroads of Cell Division, Growth and Differentiation. Nat. Rev. Cancer 2008, 8, 83–93. [Google Scholar] [CrossRef]
- Hao, B.; Oehlmann, S.; Sowa, M.E.; Harper, J.W.; Pavletich, N.P. Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases. Mol. Cell 2007, 26, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Sameer, A.S. Colorectal Cancer: Molecular Mutations and Polymorphisms. Front. Oncol. 2013, 3, 114. [Google Scholar] [CrossRef]
- Byeon, I.-J.L.; Li, H.; Song, H.; Gronenborn, A.M.; Tsai, M.-D. Sequential Phosphorylation and Multisite Interactions Characterize Specific Target Recognition by the FHA Domain of Ki67. Nat. Struct. Mol. Biol. 2005, 12, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Hu, X.; Guo, Q.; Shi, Y.; Liu, L.; Ying, G. Discovery and Validation of a Novel Metastasis-Related LncRNA Prognostic Signature for Colorectal Cancer. Front. Genet. 2022, 13, 704988. [Google Scholar] [CrossRef] [PubMed]
- Picard, N.; Caron, V.; Bilodeau, S.; Sanchez, M.; Mascle, X.; Aubry, M.; Tremblay, A. Identification of Estrogen Receptor β as a SUMO-1 Target Reveals a Novel Phosphorylated Sumoylation Motif and Regulation by Glycogen Synthase Kinase 3β. Mol. Cell. Biol. 2012, 32, 2709–2721. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, R.; Shi, W.; Huang, Z. Discovery of the Anti-Tumor Mechanism of Calycosin Against Colorectal Cancer by Using System Pharmacology Approach. Med. Sci. Monit. 2019, 25, 5589–5593. [Google Scholar] [CrossRef]
- Hayakawa, M.; Matsushima, M.; Hagiwara, H.; Oshima, T.; Fujino, T.; Ando, K.; Kikugawa, K.; Tanaka, H.; Miyazawa, K.; Kitagawa, M. Novel Insights into FGD3, a Putative GEF for Cdc42, That Undergoes SCF(FWD1/Beta-TrCP)-Mediated Proteasomal Degradation Analogous to That of Its Homologue FGD1 but Regulates Cell Morphology and Motility Differently from FGD1. Genes Cells 2008, 13, 329–342. [Google Scholar] [CrossRef] [PubMed]
- El-Masry, O.S.; Goja, A.; Rateb, M.; Owaidah, A.Y.; Alsamman, K. RNA Sequencing Identified Novel Target Genes for Adansonia Digitata in Breast and Colon Cancer Cells. Sci. Prog. 2021, 104, 368504211032084. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, G.; Alarcón, C.; Spagnoli, F.M.; Brivanlou, A.H.; Massagué, J. Balancing BMP Signaling through Integrated Inputs into the Smad1 Linker. Mol. Cell 2007, 25, 441–454. [Google Scholar] [CrossRef]
- Chu, B.; Soncin, F.; Price, B.D.; Stevenson, M.A.; Calderwood, S.K. Sequential Phosphorylation by Mitogen-Activated Protein Kinase and Glycogen Synthase Kinase 3 Represses Transcriptional Activation by Heat Shock Factor-1. J. Biol. Chem. 1996, 271, 30847–30857. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Zhang, H.; Deng, T.; Liu, R.; Liu, Y.; Li, H.; Bai, M.; Ning, T.; Wang, J.; et al. The HSF1/MiR-135b-5p Axis Induces Protective Autophagy to Promote Oxaliplatin Resistance through the MUL1/ULK1 Pathway in Colorectal Cancer. Oncogene 2021, 40, 4695–4708. [Google Scholar] [CrossRef]
- Hayakawa, M.; Kitagawa, H.; Miyazawa, K.; Kitagawa, M.; Kikugawa, K. The FWD1/Beta-TrCP-Mediated Degradation Pathway Establishes a “turning off Switch” of a Cdc42 Guanine Nucleotide Exchange Factor, FGD1. Genes Cells 2005, 10, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhao, J.; Yin, F.; Sharen, G.; Wu, Q.; Chen, Q.; Sun, X.; Yang, J.; Wang, H.; Zhang, D. A Methylation-Driven Gene Panel Predicts Survival in Patients with Colon Cancer. FEBS Open. Biol. 2021, 11, 2490–2506. [Google Scholar] [CrossRef]
- Ershov, P.; Poyarkov, S.; Konstantinova, Y.; Veselovsky, E.; Makarova, A. Transcriptomic Signatures in Colorectal Cancer Progression. Curr. Mol. Med. 2022, 23, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Fei, E.; Jia, N.; Zhang, T.; Ma, X.; Wang, H.; Liu, C.; Zhang, W.; Ding, L.; Nukina, N.; Wang, G. Phosphorylation of Ataxin-3 by Glycogen Synthase Kinase 3beta at Serine 256 Regulates the Aggregation of Ataxin-3. Biochem. Biophys. Res. Commun. 2007, 357, 487–492. [Google Scholar] [CrossRef]
- Li, D.; Zhang, T.; Lai, J.; Zhang, J.; Wang, T.; Ling, Y.; He, S.; Hu, Z. MicroRNA-25/ATXN3 Interaction Regulates Human Colon Cancer Cell Growth and Migration. Mol. Med. Rep. 2019, 19, 4213–4221. [Google Scholar] [CrossRef]
- Hart, M.; Concordet, J.P.; Lassot, I.; Albert, I.; del los Santos, R.; Durand, H.; Perret, C.; Rubinfeld, B.; Margottin, F.; Benarous, R.; et al. The F-Box Protein Beta-TrCP Associates with Phosphorylated Beta-Catenin and Regulates Its Activity in the Cell. Curr. Biol. 1999, 9, 207–210. [Google Scholar] [CrossRef]
- Liu, C.; Li, Y.; Semenov, M.; Han, C.; Baeg, G.H.; Tan, Y.; Zhang, Z.; Lin, X.; He, X. Control of Beta-Catenin Phosphorylation/Degradation by a Dual-Kinase Mechanism. Cell 2002, 108, 837–847. [Google Scholar] [CrossRef]
- Su, Y.; Fu, C.; Ishikawa, S.; Stella, A.; Kojima, M.; Shitoh, K.; Schreiber, E.M.; Day, B.W.; Liu, B. APC Is Essential for Targeting Phosphorylated Beta-Catenin to the SCFbeta-TrCP Ubiquitin Ligase. Mol. Cell 2008, 32, 652–661. [Google Scholar] [CrossRef]
- Boyle, W.J.; Smeal, T.; Defize, L.H.; Angel, P.; Woodgett, J.R.; Karin, M.; Hunter, T. Activation of Protein Kinase C Decreases Phosphorylation of C-Jun at Sites That Negatively Regulate Its DNA-Binding Activity. Cell 1991, 64, 573–584. [Google Scholar] [CrossRef]
- Turenne, G.A.; Price, B.D. Glycogen Synthase Kinase3 Beta Phosphorylates Serine 33 of P53 and Activates P53’s Transcriptional Activity. BMC Cell Biol. 2001, 2, 12. [Google Scholar] [CrossRef]
- Gregory, M.A.; Qi, Y.; Hann, S.R. Phosphorylation by Glycogen Synthase Kinase-3 Controls c-Myc Proteolysis and Subnuclear Localization. J. Biol. Chem. 2003, 278, 51606–51612. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Peng, D.; Cheng, Y. Significant Position of C-Myc in Colorectal Cancer: A Promising Therapeutic Target. Clin. Transl. Oncol. 2022, 24, 2295–2304. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.-C. Dual Regulation of Snail by GSK-3beta-Mediated Phosphorylation in Control of Epithelial-Mesenchymal Transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Pumariño, C.; Collado, M.; Castillo, M.E.; Barquín, J.; Romio, E.; Larriba, M.J.; Muñoz de Mier, G.J.; Carrato, A.; de la Pinta, C.; Pena, C. SNAI1-Expressing Fibroblasts and Derived-Extracellular Matrix as Mediators of Drug Resistance in Colorectal Cancer Patients. Toxicol. Appl. Pharmacol. 2022, 450, 116171. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C.M.; Heist, E.K.; Beals, C.R.; Crabtree, G.R.; Gardner, P. Protein Kinase A Negatively Modulates the Nuclear Accumulation of NF-ATc1 by Priming for Subsequent Phosphorylation by Glycogen Synthase Kinase-3. J. Biol. Chem. 2002, 277, 48664–48676. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Yue, C.; Wang, X.; Wang, Z.; Wu, Y.; Zhao, C.; Chang, P.; Sun, X.; Wang, W. NFATc1 Promotes Epithelial-Mesenchymal Transition and Facilitates Colorectal Cancer Metastasis by Targeting SNAI1. Exp. Cell Res. 2021, 408, 112854. [Google Scholar] [CrossRef]
- Jeong, W.-J.; Ro, E.J.; Choi, K.-Y. Interaction between Wnt/β-Catenin and RAS-ERK Pathways and an Anti-Cancer Strategy via Degradations of β-Catenin and RAS by Targeting the Wnt/β-Catenin Pathway. NPJ Precis. Oncol. 2018, 2, 5. [Google Scholar] [CrossRef]
- Nicholson, R.I.; Gee, J.M.; Harper, M.E. EGFR and Cancer Prognosis. Eur J. Cancer 2001, 37 (Suppl. 4), S9–S15. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.Y.; Richardson, B.C. The MAPK Signalling Pathways and Colorectal Cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- Ahmad, R.; Singh, J.K.; Wunnava, A.; Al-Obeed, O.; Abdulla, M.; Srivastava, S.K. Emerging Trends in Colorectal Cancer: Dysregulated Signaling Pathways (Review). Int. J. Mol. Med. 2021, 47, 14. [Google Scholar] [CrossRef]
- Mizukami, T.; Izawa, N.; Nakajima, T.E.; Sunakawa, Y. Targeting EGFR and RAS/RAF Signaling in the Treatment of Metastatic Colorectal Cancer: From Current Treatment Strategies to Future Perspectives. Drugs 2019, 79, 633–645. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Q.; Mao, J.; Cheng, L.; Shi, X.; Yu, L.; Hu, J.; Yang, M.; Li, L.; Liu, B.; et al. A Dual-Targeted Molecular Therapy of PP242 and Cetuximab Plays an Anti-Tumor Effect through EGFR Downstream Signaling Pathways in Colorectal Cancer. J. Gastrointestig. Oncol. 2021, 12, 1625–1642. [Google Scholar] [CrossRef]
- Tanoue, T.; Yamamoto, T.; Nishida, E. Modular Structure of a Docking Surface on MAPK Phosphatases. J. Biol. Chem. 2002, 277, 22942–22949. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, Y.; Steller, H. Live to Die Another Way: Modes of Programmed Cell Death and the Signals Emanating from Dying Cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, J. Role of Apoptosis in Colon Cancer Biology, Therapy, and Prevention. Curr. Colorectal Cancer Rep. 2013, 9, 331–340. [Google Scholar] [CrossRef]
- Montero, J.; Letai, A. Why Do BCL-2 Inhibitors Work and Where Should We Use Them in the Clinic? Cell Death Differ. 2018, 25, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Aouacheria, A.; Rech de Laval, V.; Combet, C.; Hardwick, J.M. Evolution of Bcl-2 Homology Motifs: Homology versus Homoplasy. Trends Cell Biol. 2013, 23, 103–111. [Google Scholar] [CrossRef]
- Letai, A.; Bassik, M.C.; Walensky, L.D.; Sorcinelli, M.D.; Weiler, S.; Korsmeyer, S.J. Distinct BH3 Domains Either Sensitize or Activate Mitochondrial Apoptosis, Serving as Prototype Cancer Therapeutics. Cancer Cell 2002, 2, 183–192. [Google Scholar] [CrossRef]
- Sanese, P.; Fasano, C.; Buscemi, G.; Bottino, C.; Corbetta, S.; Fabini, E.; Silvestri, V.; Valentini, V.; Disciglio, V.; Forte, G.; et al. Targeting SMYD3 to Sensitize Homologous Recombination-Proficient Tumors to PARP-Mediated Synthetic Lethality. iScience 2020, 23, 101604. [Google Scholar] [CrossRef]
- Fasano, C.; Lepore Signorile, M.; De Marco, K.; Forte, G.; Sanese, P.; Grossi, V.; Simone, C. Identifying Novel SMYD3 Interactors on the Trail of Cancer Hallmarks. Comput. Struct. Biotechnol. J. 2022, 20, 1860–1875. [Google Scholar] [CrossRef]
- Bottino, C.; Peserico, A.; Simone, C.; Caretti, G. SMYD3: An Oncogenic Driver Targeting Epigenetic Regulation and Signaling Pathways. Cancers 2020, 12, 142. [Google Scholar] [CrossRef] [PubMed]
- Stefanucci, A.; Luisi, G.; Zengin, G.; Macedonio, G.; Dimmito, M.P.; Novellino, E.; Mollica, A. Discovery of Arginine-Containing Tripeptides as a New Class of Pancreatic Lipase Inhibitors. Future Med. Chem. 2019, 11, 5–19. [Google Scholar] [CrossRef] [PubMed]
- McInnes, C.; Estes, K.; Baxter, M.; Yang, Z.; Farag, D.B.; Johnston, P.; Lazo, J.S.; Wang, J.; Wyatt, M.D. Targeting Subcellular Localization through the Polo-Box Domain: Non-ATP Competitive Inhibitors Recapitulate a PLK1 Phenotype. Mol. Cancer Ther. 2012, 11, 1683–1692. [Google Scholar] [CrossRef] [PubMed]
- Milorey, B.; Schweitzer-Stenner, R.; Andrews, B.; Schwalbe, H.; Urbanc, B. Short Peptides as Predictors for the Structure of Polyarginine Sequences in Disordered Proteins. Biophys. J. 2021, 120, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Su, A.; Tabata, Y.; Aoki, K.; Sada, A.; Ohki, R.; Nagatoishi, S.; Tsumoto, K.; Wang, S.; Otani, Y.; Ohwada, T. Elaboration of Non-Naturally Occurring Helical Tripeptides as P53-MDM2/MDMX Interaction Inhibitors. Chem. Pharm. Bull. (Tokyo) 2021, 69, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Trost, B.; Kanduc, D.; Kusalik, A. Rare Peptide Segments Are Found Significantly More Often in Proto-Oncoproteins than Control Proteins: Implications for Immunology and Oncology. J. R. Soc. Interface 2009, 6, 123–127. [Google Scholar] [CrossRef]
- Kusalik, A.; Trost, B.; Bickis, M.; Fasano, C.; Capone, G.; Kanduc, D. Codon Number Shapes Peptide Redundancy in the Universal Proteome Composition. Peptides 2009, 30, 1940–1944. [Google Scholar] [CrossRef]
- Kanduc, D. Protein Information Content Resides in Rare Peptide Segments. Peptides 2010, 31, 983–988. [Google Scholar] [CrossRef]
- Ahn, Y.-H.; Chang, Y.-T. Tagged Small Molecule Library Approach for Facilitated Chemical Genetics. Acc. Chem. Res. 2007, 40, 1025–1033. [Google Scholar] [CrossRef]
- Cisar, J.S.; Cravatt, B.F. Fully Functionalized Small-Molecule Probes for Integrated Phenotypic Screening and Target Identification. J. Am. Chem. Soc. 2012, 134, 10385–10388. [Google Scholar] [CrossRef]
- Kambe, T.; Correia, B.E.; Niphakis, M.J.; Cravatt, B.F. Mapping the Protein Interaction Landscape for Fully Functionalized Small-Molecule Probes in Human Cells. J. Am. Chem. Soc. 2014, 136, 10777–10782. [Google Scholar] [CrossRef]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 615056. [Google Scholar] [CrossRef]
- Via, A.; Uyar, B.; Brun, C.; Zanzoni, A. How Pathogens Use Linear Motifs to Perturb Host Cell Networks. Trends Biochem. Sci. 2015, 40, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Sámano-Sánchez, H.; Gibson, T.J. Mimicry of Short Linear Motifs by Bacterial Pathogens: A Drugging Opportunity. Trends Biochem. Sci. 2020, 45, 526–544. [Google Scholar] [CrossRef] [PubMed]
- Zanzoni, A.; Spinelli, L.; Braham, S.; Brun, C. Perturbed Human Sub-Networks by Fusobacterium Nucleatum Candidate Virulence Proteins. Microbiome 2017, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Schuster-Böckler, B.; Bateman, A. Protein Interactions in Human Genetic Diseases. Genome Biol. 2008, 9, R9. [Google Scholar] [CrossRef] [PubMed]
- Teng, S.; Madej, T.; Panchenko, A.; Alexov, E. Modeling Effects of Human Single Nucleotide Polymorphisms on Protein-Protein Interactions. Biophys. J. 2009, 96, 2178–2188. [Google Scholar] [CrossRef]
- Whitty, A.; Kumaravel, G. Between a Rock and a Hard Place? Nat. Chem. Biol. 2006, 2, 112–118. [Google Scholar] [CrossRef]
- Arkin, M.R.; Wells, J.A. Small-Molecule Inhibitors of Protein-Protein Interactions: Progressing towards the Dream. Nat. Rev. Drug Discov. 2004, 3, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Higueruelo, A.P.; Schreyer, A.; Bickerton, G.R.J.; Pitt, W.R.; Groom, C.R.; Blundell, T.L. Atomic Interactions and Profile of Small Molecules Disrupting Protein-Protein Interfaces: The TIMBAL Database. Chem. Biol. Drug Des. 2009, 74, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Morell, M.; Avilés, F.X.; Ventura, S. Detecting and Interfering Protein Interactions: Towards the Control of Biochemical Pathways. Curr. Med. Chem. 2009, 16, 362–379. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J. Inhibition of Protein-Protein Interactions Using Designed Molecules. Chem. Soc. Rev. 2009, 38, 3289–3300. [Google Scholar] [CrossRef] [PubMed]
- Peserico, A.; Germani, A.; Sanese, P.; Barbosa, A.J.; Di Virgilio, V.; Fittipaldi, R.; Fabini, E.; Bertucci, C.; Varchi, G.; Moyer, M.P.; et al. A SMYD3 Small-Molecule Inhibitor Impairing Cancer Cell Growth. J. Cell. Physiol. 2015, 230, 2447–2460. [Google Scholar] [CrossRef] [PubMed]
- Van Aller, G.S.; Graves, A.P.; Elkins, P.A.; Bonnette, W.G.; McDevitt, P.J.; Zappacosta, F.; Annan, R.S.; Dean, T.W.; Su, D.-S.; Carpenter, C.L.; et al. Structure-Based Design of a Novel SMYD3 Inhibitor That Bridges the SAM-and MEKK2-Binding Pockets. Structure 2016, 24, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.H.; Boriack-Sjodin, P.A.; Smith, S.; Thomenius, M.; Rioux, N.; Munchhof, M.; Mills, J.E.; Klaus, C.; Totman, J.; Riera, T.V.; et al. Novel Oxindole Sulfonamides and Sulfamides: EPZ031686, the First Orally Bioavailable Small Molecule SMYD3 Inhibitor. ACS Med. Chem. Lett. 2016, 7, 134–138. [Google Scholar] [CrossRef]
- Jain, A.K.; Jain, S. Advances in Oral Delivery of Anti-Cancer Prodrugs. Expert Opin. Drug Deliv. 2016, 13, 1759–1775. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.B.; Ganesan, A.; Emery, F.S. Oral Administration of Peptide-Based Drugs: Beyond Lipinski’s Rule. ChemMedChem 2016, 11, 2245–2251. [Google Scholar] [CrossRef]
- Borghouts, C.; Kunz, C.; Delis, N.; Groner, B. Monomeric Recombinant Peptide Aptamers Are Required for Efficient Intracellular Uptake and Target Inhibition. Mol. Cancer Res. 2008, 6, 267–281. [Google Scholar] [CrossRef]
- Borghouts, C.; Tittmann, H.; Delis, N.; Kirchenbauer, M.; Brill, B.; Groner, B. The Intracellular Delivery of a Recombinant Peptide Derived from the Acidic Domain of PIAS3 Inhibits STAT3 Transactivation and Induces Tumor Cell Death. Mol. Cancer Res. 2010, 8, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.; Brill, B.; Borghouts, C.; Delis, N.; Mack, L.; Groner, B. Survivin Inhibition by an Interacting Recombinant Peptide, Derived from the Human Ferritin Heavy Chain, Impedes Tumor Cell Growth. J. Cancer Res. Clin. Oncol. 2012, 138, 1205–1220. [Google Scholar] [CrossRef]
- Kadkhodayan, S.; Elliott, L.O.; Mausisa, G.; Wallweber, H.A.; Deshayes, K.; Feng, B.; Fairbrother, W.J. Evaluation of Assay Technologies for the Identification of Protein-Peptide Interaction Antagonists. Assay Drug Dev. Technol. 2007, 5, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Uehara, Y.; Mochizuki, M.; Matsuno, K.; Haino, T.; Asai, A. Novel High-Throughput Screening System for Identifying STAT3-SH2 Antagonists. Biochem. Biophys. Res. Commun. 2009, 380, 627–631. [Google Scholar] [CrossRef]
- Murray, E.J.; Leaman, D.P.; Pawa, N.; Perkins, H.; Pickford, C.; Perros, M.; Zwick, M.B.; Butler, S.L. A Low-Molecular-Weight Entry Inhibitor of Both CCR5- and CXCR4-Tropic Strains of Human Immunodeficiency Virus Type 1 Targets a Novel Site on Gp41. J. Virol. 2010, 84, 7288–7299. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.A.; Kozhevnikova, M.V.; Cexus, O.N.F.; Zamyatnin, A.A.; Soond, S.M. BH3-Mimetics: Recent Developments in Cancer Therapy. J. Exp. Clin. Cancer Res. 2021, 40, 355. [Google Scholar] [CrossRef] [PubMed]
- Huerta, S.; Gao, X.; Livingston, E.H.; Kapur, P.; Sun, H.; Anthony, T. In Vitro and in Vivo Radiosensitization of Colorectal Cancer HT-29 Cells by the Smac Mimetic JP-1201. Surgery 2010, 148, 346–353. [Google Scholar] [CrossRef]
- Ohba, M.; Mizuki, E.; Uemori, A. Parasporin, a New Anticancer Protein Group from Bacillus Thuringiensis. Anticancer Res. 2009, 29, 427–433. [Google Scholar]
- Kitada, S.; Abe, Y.; Shimada, H.; Kusaka, Y.; Matsuo, Y.; Katayama, H.; Okumura, S.; Akao, T.; Mizuki, E.; Kuge, O.; et al. Cytocidal Actions of Parasporin-2, an Anti-Tumor Crystal Toxin from Bacillus Thuringiensis. J. Biol. Chem. 2006, 281, 26350–26360. [Google Scholar] [CrossRef]
- Cruz, J.; Suárez-Barrera, M.O.; Rondón-Villarreal, P.; Olarte-Diaz, A.; Guzmán, F.; Visser, L.; Rueda-Forero, N.J. Computational Study, Synthesis and Evaluation of Active Peptides Derived from Parasporin-2 and Spike Protein from Alphacoronavirus against Colorectal Cancer Cells. Biosci. Rep. 2021, 41, BSR20211964. [Google Scholar] [CrossRef] [PubMed]
- Akiba, T.; Okumura, S. Parasporins 1 and 2: Their Structure and Activity. J. Invertebr. Pathol. 2017, 142, 44–49. [Google Scholar] [CrossRef]
- Mizuki, E.; Park, Y.S.; Saitoh, H.; Yamashita, S.; Akao, T.; Higuchi, K.; Ohba, M. Parasporin, a Human Leukemic Cell-Recognizing Parasporal Protein of Bacillus Thuringiensis. Clin. Diagn. Lab. Immunol. 2000, 7, 625–634. [Google Scholar] [CrossRef]
- Brasseur, K.; Auger, P.; Asselin, E.; Parent, S.; Côté, J.-C.; Sirois, M. Parasporin-2 from a New Bacillus Thuringiensis 4R2 Strain Induces Caspases Activation and Apoptosis in Human Cancer Cells. PLoS ONE 2015, 10, e0135106. [Google Scholar] [CrossRef]
- Reich, N.C.; Liu, L. Tracking STAT Nuclear Traffic. Nat. Rev. Immunol. 2006, 6, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Deng, J.; Kujawski, M.; Yang, C.; Liu, Y.; Herrmann, A.; Kortylewski, M.; Horne, D.; Somlo, G.; Forman, S.; et al. STAT3-Induced S1PR1 Expression Is Crucial for Persistent STAT3 Activation in Tumors. Nat. Med. 2010, 16, 1421–1428. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-H.; Wei, H.; Lv, S.-Q.; Ji, H.; Wang, D.-L. Knockdown of STAT3 Expression by RNAi Suppresses Growth and Induces Apoptosis and Differentiation in Glioblastoma Stem Cells. Int. J. Oncol. 2010, 37, 103–110. [Google Scholar] [PubMed]
- Norouzi, P.; Mirmohammadi, M.; Houshdar Tehrani, M.H. Anticancer Peptides Mechanisms, Simple and Complex. Chem. Biol. Interact 2022, 368, 110194. [Google Scholar] [CrossRef] [PubMed]
- Chantawannakul, J.; Chatpattanasiri, P.; Wattayagorn, V.; Kongsema, M.; Noikaew, T.; Chumnanpuen, P. Virtual Screening for Biomimetic Anti-Cancer Peptides from Cordyceps Militaris Putative Pepsinized Peptidome and Validation on Colon Cancer Cell Line. Molecules 2021, 26, 5767. [Google Scholar] [CrossRef] [PubMed]
- Hadianamrei, R.; Tomeh, M.A.; Brown, S.; Wang, J.; Zhao, X. Rationally Designed Short Cationic α-Helical Peptides with Selective Anticancer Activity. J. Colloid Interface Sci. 2022, 607, 488–501. [Google Scholar] [CrossRef] [PubMed]
- Cerella, C.; Dicato, M.; Diederich, M. BH3 Mimetics in AML Therapy: Death and Beyond? Trends Pharmacol. Sci. 2020, 41, 793–814. [Google Scholar] [CrossRef]
- Hao, Y.; Chung, C.K.; Gu, Z.; Schomann, T.; Dong, X.; Veld, R.V.H.; Camps, M.G.M.; Ten Dijke, P.; Ossendorp, F.A.; Cruz, L.J. Combinatorial Therapeutic Approaches of Photodynamic Therapy and Immune Checkpoint Blockade for Colon Cancer Treatment. Mol. Biomed. 2022, 3, 26. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Verhagen, A.M.; Lock, P. Revealing the Intricacies of Cancer. Genome Biol. 2002, 3, reports4015.1. [Google Scholar] [CrossRef] [PubMed]

| Name of In Silico Resource | Website | Description | Technical Advantages | Refs |
|---|---|---|---|---|
| PINA (Protein Interaction Network Analysis) | https://omics.bjcancer.org/pina/ (accessed on 3 September 2022) | An integrated server for PPI non-redundant and curated data referred to six model organisms | A useful tool to provide comprehensive PPI information through integrated visualization of PPI data and construction, filtering, and data analysis | [40,41] |
| SPRINT (Scoring Protein INTeractions) | www.csd.uwo.ca/faculty/ilie/SPRINT (accessed on 3 September 2022) | This algorithm enables the computational organization of existing PPI networks to the level of species interactomes | It simplifies the interpretation of the results and makes them more objective through a PPI score | [42] |
| Path2PPI | http://bioconductor.org/packages/release/bioc/html/Path2PPI.html (accessed on 3 September 2022) | This algorithm analyzes the homology between protein sequences of multiple organisms or a single target organism. | It allows users to combine sequence similarity searches of the examined proteins with their functional information about a particular pathway | [43] |
| Paralog Matching | https://github.com/Mirmu/ParalogMatching.jl (accessed on 3 September 2022) | This algorithm predicts interacting paralogs between two distinct protein families. It enables homology analysis of all members of two protein families that belong to the same species and predicts PPIs, maximizing the detectable coevolutionary signal. | It provides a direct correlation by amino acid occurrences between multiple sequence alignment (MSA) and interprotein residue–residue contacts in the PPI | [44] |
| RaptorX-ComplexContact server | http://raptorx.uchicago.edu/ (accessed on 3 September 2022) | This server analyzes the interfacial contacts between two potentially interacting heterodimeric protein sequences using deep-learning techniques. | A useful tool for protein docking analysis, protein–protein interaction prediction, and protein interaction network construction | [45] |
| COZOID (Contact Zone Identifier) | http://decibel.fi.muni.cz/cozoid (accessed on 3 September 2022) | This algorithm analyzes several docking structures covering the three major types of PPIs (coiled-coil, pocket-string, and surface–surface interactions) and their contact zones with different levels of detail. | It provides docking models of interacting proteins and enables the selection of the best docking structures based on their similarity to a conserved structure from reference homologous proteins | [46] |
| Path-LZerD | https://kiharalab.org/proteindocking/pathlzerd.php (accessed on 3 September 2022) | This software predicts the assembly order of multimeric proteins starting from single subunit structures. | A useful tool to design drugs that target crucial interactions within a specific complex | [47] |
| ReactomeFIViz | https://reactome.org/tools/reactome-fiviz (accessed on 3 September 2022) | This Cytoscape application facilitates the pathway- and network-based analysis of RNA-seq and other omics datasets using the Reactome pathway database. | It allows users to link the PPI dataset reported in two or more databases | [48] |
| KeyPathwayMiner | https://apps.cytoscape.org/apps/keypathwayminer (accessed on 3 September 2022) | This Cytoscape application detects highly connected PPI networks in which genes show similar expression. | A useful tool to combine interaction network data with omics datasets in order to identify novel functional peptide modules | [49] |
| VieClus (Vienna Graph Clustering) | http://vieclus.taa.univie.ac.at/ (accessed on 3 September 2022) | This software enables the visualization of PPI clusters showing similar functional modules. | It allows users to identify functional modules by searching for sets of proteins whose interactions are dense within the sets but sparse between the sets | [50] |
| TD-WGcluster (Time Delayed Weighted Edge Clustering portal) | https://www.r-project.org/ (accessed on 3 September 2022) | This algorithm integrates the three-dimensional topology of PPIs. | It combines PPI topology with a dynamics component derived from time series data | [51] |
| SANA (Simulated Annealing Network Aligner) | https://sana.ics.uci.edu/ (accessed on 3 September 2022) | This alignment software compares PPI motifs between different species. | A useful tool to perform comparative analyses of PPI networks to reveal evolutionary relationships between species | [52] |
| PEPPI (Predicted Protein-protein Interactions) | https://zhanggroup.org›PEPPI (accessed on 3 September 2022) | This alignment software predicts the exact peptide modules involved in binary interactions between two amino acid sequences. | It integrates multiple independent prediction and analysis methods of protein sequence similarity, structural homology, functional association, and machine learning-based classification | [53] |
| CPDB (Consensus PathDB) | http://cpdb.molgen.mpg.de/ (accessed on 3 September 2022) | A comprehensive database for studying human PPI networks and related information (biochemical pathway, genetic, metabolic, signaling data, and drug–target interactions). | A useful tool to provide a correct interpretation of the massive quantities of PPI molecular data | [54] |
| IID (Integrated Interactions Database) | http://iid.ophid.utoronto.ca/ (accessed on 3 September 2022) | A curated database containing comprehensive information on PPIs detected and predicted in 18 species, including humans. | A useful tool to study PPIs in specific conditions (e.g., tissues, developmental stages), conservation across species, directionality of the interaction, and druggability | [55] |
| HIPPIE (Human Integrated Protein-Protein Interaction rEference) | http://cbdm-01.zdv.uni-mainz.de/~mschaefer/hippie/ (accessed on 3 September 2022) | This resource integrates multiple human PPI databases. | It allows users to overlay gene expression data and other annotation resources to construct protein networks specific to a tissue, disease, or subcellular localization | [56] |
| MIPS (Mammalian Protein-Protein Interaction Database) | http://mips.helmholtz-muenchen.de/proj/ppi/ (accessed on 3 September 2022) | A manually curated database of high-quality PPI data from the scientific literature. | A useful tool for the metanalysis of current scientific literature on mammalian PPIs | [57] |
| OncoPPi Portal | https://oncoppi.emory.edu/ (accessed on 3 September 2022) | A comprehensive PPI network database concerning cancer; | This tool is used in cancer research to provide genetic, pharmacological, clinical, and structural data and combine them with the network of cancer-associated PPIs experimentally found in tumor cells | [38,39] |
| BioGRID | https://thebiogrid.org/ (accessed on 3 September 2022) | A comprehensive repository of PPI data providing information on their druggability; | A useful tool to study oncoprotein-chemical compound associations, based on experimental data (freely available in a variety of standardized formats) | [58] |
| IntAct | https://www.ebi.ac.uk/intact/home (accessed on 3 September 2022) | This is an open-source PPI database. IntAct data are organized in three clusters of information (proteomes, datasets, and mutations) that simplify the search for database entries. | A useful tool to analyze current experimentally derived PPI data from the published scientific literature; it also offers free tools for integration and analysis purposes | [59] |
| Name of In Silico Resource | Website | Description | Technical Advantages | Refs |
|---|---|---|---|---|
| PIPE (Protein–Protein Interaction Prediction Engine) | https://pipe.rcc.fsu.edu/ (accessed on 4 September 2022) | This algorithm predicts the binding sites involved in PPIs based on query protein sequences and a database of known binary interaction data. The outcome is a three-dimensional graph where the peaks signify a high co-occurrence of the corresponding sequences among known interacting proteins | A useful tool to identify PPI consensus motifs. The PIPE method is based on re-occurrences of peptide sequences that mediate a large number of PPIs. | [62] |
| MnM (Minimotif Miner database) | http://minimotifminer.org or http://mnm.engr.uconn.edu (accessed on 4 September 2022) | This comprehensive database reports over 300,000 functional SLiMs in protein queries. | A database designed to improve prediction accuracy, allowing users to search for SLiMs with a set of false-positive filters and linear regression scoring. | [63] |
| LMPID (Linear Motif Mediated Protein Interaction Database) | http://bicresources.jcbose.ac.in/ssaha4/lmpid (accessed on 4 September 2022) | This manually curated database reports experimentally validated data about SLiM-mediated PPIs from any organism. It contains comprehensive information about 1762 unique SLiMs mediating 2215 PPIs among 1187 bait and 559 prey proteins. | This tool is mainly used to improve knowledge of the patterns of SLiMs binding to a specific domain and to formulate PPI inhibitors/modulators of interest. | [64] |
| ADAN (protein-protein interAction of moDular domAiN database) | https://adan-embl.ibmc.umh.es/ (accessed on 4 September 2022) | This manually integrated and curated database is used for the prediction of PPI-mediating SLiMs. It currently contains 3505 entries comprising structural and functional SLiM information (biochemical data, sequence files, and alignments), which is cross-referenced to other databases. The in silico prediction method is based on position-specific scoring matrices. | A useful tool to predict exact SLiMs and the best ligand and putative binding partner candidates of a protein of interest. | [65] |
| ELM (Eukaryotic Linear Motif) | http://elm.eu.org/ (accessed on 4 September 2022) | This manually curated platform contains different types of experimentally validated SLiM data from current literature. The classification of ELM entries is based on motif type, functional site, and ELM class. The ELM class is a specific list of experimentally validated SLiMs matching the examined query sequence. | A very versatile resource that is useful for various purposes in SLiM-related studies. It provides both a database of annotated SLiM data and an exploratory tool to predict them. | [15] |
| SLiMAN (is a recent database) | https://sliman.cbs.cnrs.fr (accessed on 4 September 2022) | This web server contains complementary information from the Uniprot (https://www.uniprot.org/; accessed on 4 September 2022), ELM (http://elm.eu.org/; accessed on 4 September 2022), IUpred2 (https://iupred2a.elte.hu/; accessed on 4 September 2022), BioGrid (https://thebiogrid.org/; accessed on 4 September 2022), and PhosphoSitePlus (https://www.phosphosite.org/; accessed on 4 September 2022) databases. These databases have been integrated to provide a comprehensive analysis of SLiM sequences (annotated in the ELM database), motif disorder scores (annotated in the IUpred2 server), predicted and experimentally validated PPIs (annotated in the BioGrid database), and PTMs (annotated in the Uniprot and PhosphoSitePlus databases). | A useful tool designed to overcome the various limitations related to the complex characteristics intrinsic to SLiMs (i.e., their typical localization in disordered regions or loops, their short but variable length, the varying conservation of their sequence, and their slightly bent structure). | [60] |
| Uniprot Acc. #, Gene, Entry Name | SLiM Start | SLiM End | SLiM Sequence | No. of Evidence | Experimental Evidence | Refs |
|---|---|---|---|---|---|---|
| Q9BYG3, MKI67IP MK67I_HUMAN | 227 231 | 234 238 | LDTPEKTVDSQGPTPVCTPT EKTVDSQGPTPVCTPTFLER | 3 3 | Protein kinase assay; mass spectrometry; mutation analysis | [86,87] |
| Q92731, ESR2 ESR2_HUMAN | 5 | 12 | MDIKNSPSSLNSPSSYNCSQ | 3 | Inhibitor; western blotting; mutation analysis; | [88,89] |
| Q5JSP0, FGD3 FGD3_HUMAN | 73 77 | 80 84 | GSLKIPNRDSGIDSPSSSVA IPNRDSGIDSPSSSVAGENF | 5 2 | Protein kinase assay; mutation analysis; co-immunoprecipitation; alanine scanning | [90,91] |
| Q15797, SMAD1 SMAD1_HUMAN | 199 207 | 206 214 | PNSPGSSSSTYPHSPTSSDP STYPHSPTSSDPGSPFQMPA | 4 4 | Protein kinase assay; radiolabeling; mutation analysis; western blotting | [92] |
| Q00613, HSF1 HSF1_HUMAN | 300 | 307 | LVRVKEEPPSPPQSPRVEEA | 2 | Protein kinase assay; mutation analysis; | [93,94] |
| P98174, FGD1 FGD1_HUMAN | 280 | 287 | DGEKVPNRDSGIDSISSPSN | 1 | Inhibitor | [95,96] |
| P84022, SMAD3 SMAD3_HUMAN | 201 | 208 | QMNHSMDAGSPNLSPNPMSP | 3 | Knock out; mutation analysis; protein kinase assay | [72,97] |
| P54252 ATXN3 ATX3_HUMAN | 253 | 260 | ADLRRAIQLSMQGSSRNISQ | 2 | Protein kinase assay; mutation analysis | [98,99] |
| P35222, CTNNB1 CTNB1_HUMAN | 30 | 37 | HWQQQSYLDSGIHSGATTTA | 4 | Protein kinase assay; inhibitor | [100,101,102] |
| P24864, CCNE1 CCNE1_HUMAN | 373 388 392 | 380 395 399 | EQNRASPLPSGLLTPPQSGKEQ EQNRASPLPSGLLTPPQSGK ASPLPSGLLTPPQSGKKQSS | 4 4 1 | Protein kinase assay; mutation analysis; two-dimensional phosphopeptide mapping | [83] |
| P05412, JUN JUN_HUMAN | 236 | 243 | QTVPEMPGETPPLSPIDMES | 1 | Two-dimensional phosphopeptide mapping | [103] |
| P04637, TP53 P53_HUMAN | 30 | 37 | KLLPENNVLSPLPSQAMDDL | 2 | Protein kinase assay; mutation analysis | [104] |
| P01106, MYC MYC_HUMAN | 55 | 62 | IWKKFELLPTPPLSPSRRSG | 2 | Co-immunoprecipitation; western blotting | [105,106] |
| O95863, SNAI1 SNAI1_HUMAN | 93 | 100 | ELTSLSDEDSGKGSQPPSPP | 3 | Co-immunoprecipitation; western blotting; alanine scanning | [107,108] |
| O95644, NFATC1 NFAC1_HUMAN | 238 287 | 245 294 | GSPRHSPSTSPRASVTEESW HSPTPSPHGSPRVSVTDDSW | 2 2 | Protein kinase assay; mutation analysis | [109,110] |
| Trial Status | Trial ID | Title | Treatment (s) | Result Availability |
|---|---|---|---|---|
| Completed | NCT00019331 | Vaccine Therapy Plus Biological Therapy in Treating Adults with Metastatic Solid Tumors | Biological: Ras peptide cancer vaccine Biological: aldesleukin Biological: sargramostim Drug: DetoxPC | Not available |
| NCT00098943 | NGR-TNF in Treating Patients with Advanced Solid Tumors | Biological: CNGRC peptide-TNF alpha conjugate | Not available | |
| NCT00020267 | Vaccine Therapy in Treating Patients with Metastatic Cancer | Drug: interleukin-2 Drug: MAGE-12 peptide vaccine Drug: montanide ISA-51 | Not available | |
| NCT01364844 | Safety and Tolerability of DS-7423 in Subjects with Advanced Solid Malignant Tumors | Drug: DS-7423 | Not available | |
| NCT00841191 | A Safety, Efficacy, and Pharmacokinetic Study of Siltuximab (CNTO 328) in Participants with Solid Tumors | Drug: CNTO 328; anti-interleukin-6 monoclonal antibody | Not available | |
| Terminated | NCT00091286 | Vaccine Therapy in Treating Patients with Stage IIB, Stage III, or Stage IV Colorectal Cancer | Biological: HER-2-neu, CEA peptides, GM-CSF, montanide ISA-51 vaccine | Not available |
| NCT00677612 | Histocompatibility Leukocyte Antigen (HLA)-A*0201 Restricted Peptide Vaccine Therapy in Patients with Colorectal Cancer | Biological: VEGFR1 and VEGFR2 | Not available | |
| NCT00677287 | Histocompatibility Leukocyte Antigen (HLA)-A*2402 Restricted Peptide Vaccine Therapy in Patients with Colorectal Cancer | Biological: RNF43, TOMM34, VEGFR1 and VEGFR2 | Not available | |
| NCT02300922 | Pretargeted Radioimmunotherapy in Metastatic Colorectal Cancer | Drug: antibody TF2 Drug: 90-Y-IMP-288 Drug: 111-In-IMP-288 | Not available | |
| NCT03724253 | [68Ga]-NeoBOMB1 Imaging in Patients with Malignancies Known to Overexpress Gastrin Releasing Peptide Receptor (GRPR) | Drug: [68Ga]-NeoBOMB1 | Available | |
| NCT00012246 | Vaccine Therapy in Treating Patients with Cancer of the Gastrointestinal Tract | Biological: carcinoembryonic antigen peptide 1–6D Biological: incomplete Freund’s adjuvant Biological: sargramostim | Not available | |
| NCT01925274 | A Study Of PF-05212384 Plus Irinotecan vs. Cetuximab Plus Irinotecan in Patients with KRAS And NRAS Wild Type Metastatic Colorectal Cancer | Drug: PF-05212384 Drug: irinotecan Drug: cetuximab Drug: irinotecan | Available | |
| NCT03148119 | Study of QRH-882260 Heptapeptide Application in the Colon | Drug: QRH-882260 heptapeptide Device: scanning fiber endoscope | Not available |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fasano, C.; Grossi, V.; Forte, G.; Simone, C. Short Linear Motifs in Colorectal Cancer Interactome and Tumorigenesis. Cells 2022, 11, 3739. https://doi.org/10.3390/cells11233739
Fasano C, Grossi V, Forte G, Simone C. Short Linear Motifs in Colorectal Cancer Interactome and Tumorigenesis. Cells. 2022; 11(23):3739. https://doi.org/10.3390/cells11233739
Chicago/Turabian StyleFasano, Candida, Valentina Grossi, Giovanna Forte, and Cristiano Simone. 2022. "Short Linear Motifs in Colorectal Cancer Interactome and Tumorigenesis" Cells 11, no. 23: 3739. https://doi.org/10.3390/cells11233739