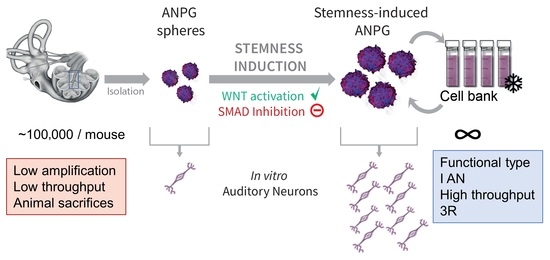
WNT Activation and TGFβ-Smad Inhibition Potentiate Stemness of Mammalian Auditory Neuroprogenitors for High-Throughput Generation of Functional Auditory Neurons In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Spiral Ganglion Cells Isolation, In Vitro Culture and Differentiation
2.2. Cell Counting
2.3. Automated Analysis of ANPGs Neurosphere Growth
2.4. Cell Cycle Analysis by Flow Cytometry (FACS)
2.5. RNA Sequencing
2.6. Video Time Lapse Microscopy
2.7. Immunofluorescence and Confocal Microscopy
2.8. Electrophysiological Characterization of Differentiated SGNs
2.9. Statistical Analysis
3. Results
3.1. Intrinsic Stemness Properties of ANPGs Isolated from C57Bl/6 and A/J Mice
3.2. Comparative Transcriptomic Analysis (RNA-Sequencing) of High- vs. Low Stemness Neuroprogenitors
3.3. Pathway Enrichment Analysis and Differentially Represented Gene Ontologies between High and Low Stemness ANPGs
3.4. Reprogramming Stemness of Presenescent ANPGs
3.5. Phenotype of Stemness-Induced ANPGs (si-ANPGs)
3.6. Electrophysiological Characterization of Stemness-Induced Auditory Neurons (si-ANs) Differentiated from si-ANPGs
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, B.S.; Tucci, D.L.; Merson, M.H.; O’Donoghue, G.M. Global hearing health care: New findings and perspectives. Lancet 2017, 390, 2503–2515. [Google Scholar] [CrossRef]
- Corwin, T.J.; Cotanche, D.A. Regeneration of sensory hair cells after acoustic trauma. Science 1988, 240, 1772–1774. [Google Scholar] [CrossRef]
- Ryals, M.B.; Rubel, E.W. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science 1988, 240, 1774–1776. [Google Scholar] [CrossRef] [PubMed]
- Balak, K.J.; Corwin, J.T.; Jones, J.E. Regenerated hair cells can originate from supporting cell progeny: Evidence from phototoxicity and laser ablation experiments in the lateral line system. J. Neurosci. 1990, 10, 2502–2512. [Google Scholar] [CrossRef] [PubMed]
- Lombarte, A.; Yan, H.Y.; Popper, A.N.; Chang, J.S.; Platt, C. Damage and regeneration of hair cell ciliary bundles in a fish ear following treatment with gentamicin. Hear. Res. 1993, 64, 166–174. [Google Scholar] [CrossRef]
- Burns, J.C.; Cox, B.C.; Thiede, B.R.; Zuo, J.; Corwin, J.T. In vivo proliferative regeneration of balance hair cells in newborn mice. J. Neurosci. 2012, 32, 6570–6577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forge, A.; Li, L.; Corwin, J.; Nevill, G. Ultrastructural evidence for hair cell regeneration in the mammalian inner ear. Science 1993, 259, 1616–1619. [Google Scholar] [CrossRef] [PubMed]
- Golub, J.S.; Tong, L.; Ngyuen, T.B.; Hume, C.R.; Palmiter, R.D.; Rubel, E.W.; Stone, J.S. Hair cell replacement in adult mouse utricles after targeted ablation of hair cells with diphtheria toxin. J. Neurosci. 2012, 32, 15093–15105. [Google Scholar] [CrossRef] [Green Version]
- Kawamoto, K.; Izumikawa, M.; Beyer, L.A.; Atkin, G.M.; Raphael, Y. Spontaneous hair cell regeneration in the mouse utricle following gentamicin ototoxicity. Hear. Res. 2009, 247, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Warchol, M.; Lambert, P.; Goldstein, B.; Forge, A.; Corwin, J. Regenerative proliferation in inner ear sensory epithelia from adult guinea pigs and humans. Science 1993, 259, 1619–1622. [Google Scholar] [CrossRef]
- Bohne, B.A.; Ward, P.H.; Fernandez, C. Irreversible inner ear damage from rock music. Trans. Sect. Otolaryngol. Am. Acad. Ophthalmol. Otolaryngol. 1976, 82, ORL50-9. [Google Scholar]
- Hawkins, J.E.; Johnsson, L.-G.; Stebbins, W.C.; Moody, D.B.; Coombs, S.L. Hearing loss and cochlear pathology in monkeys after noise exposure. Acta Otolaryngol. 1976, 81, 337–343. [Google Scholar] [CrossRef]
- Oesterle, E.C.; Campbell, S.; Taylor, R.R.; Forge, A.; Hume, C.R. Sox2 and Jagged1 Expression in Normal and Drug-Damaged Adult Mouse Inner Ear. J. Assoc. Res. Otolaryngol. 2007, 9, 65–89. [Google Scholar] [CrossRef] [Green Version]
- Rask-Andersen, H.; Boström, M.; Gerdin, B.; Kinnefors, A.; Nyberg, G.; Engstrand, T.; Miller, J.M.; Lindholm, D. Regeneration of human auditory nerve. In vitro/in video demonstration of neural progenitor cells in adult human and guinea pig spiral ganglion. Hear. Res. 2005, 203, 180–191. [Google Scholar] [CrossRef]
- Senn, P.; Mina, A.; Volkenstein, S.; Kranebitter, V.; Oshima, K.; Heller, S. Progenitor Cells from the Adult Human Inner Ear. Anat. Rec. 2019, 303, 461–470. [Google Scholar] [CrossRef]
- Oshima, K.; Grimm, C.M.; Corrales, C.E.; Senn, P.; Monedero, R.M.; Géléoc, G.S.G.; Edge, A.; Holt, J.R.; Heller, S. Differential Distribution of Stem Cells in the Auditory and Vestibular Organs of the Inner Ear. J. Assoc. Res. Otolaryngol. 2006, 8, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Moon, B.-S.; Ammothumkandy, A.; Zhang, N.; Peng, L.; Ibrayeva, A.; Bay, M.; Pratap, A.; Park, H.J.; Bonaguidi, M.A.; Lu, W. The Presence of Neural Stem Cells and Changes in Stem Cell-Like Activity With Age in Mouse Spiral Ganglion Cells In Vivo and In Vitro. Clin. Exp. Otorhinolaryngol. 2018, 11, 224–232. [Google Scholar] [CrossRef] [Green Version]
- Rousset, F.; Kokje, V.B.C.; Sipione, R.; Schmidbauer, D.; Nacher-Soler, G.; Ilmjärv, S.; Coelho, M.; Fink, S.; Voruz, F.; El Chemaly, A.; et al. Intrinsically Self-renewing Neuroprogenitors From the A/J Mouse Spiral Ganglion as Virtually Unlimited Source of Mature Auditory Neurons. Front. Cell. Neurosci. 2020, 14, 395. [Google Scholar] [CrossRef]
- Tritsch, X.N.; Bergles, D.E. Developmental regulation of spontaneous activity in the Mammalian cochlea. J. Neurosci. 2010, 30, 1539–1550. [Google Scholar] [CrossRef] [Green Version]
- Oshima, K.; Senn, P.; Heller, S. Isolation of sphere-forming stem cells from the mouse inner ear. Methods Mol. Biol. 2009, 493, 141–162. [Google Scholar]
- Rousset, F.; Schmidbauer, D.; Fink, S.; Adel, Y.; Obexer, B.; Müller, M.; Glueckert, R.; Löwenheim, H.; Senn, P. Phoenix auditory neurons as 3R cell model for high throughput screening of neurogenic compounds. Hear. Res. 2021, 414, 108391. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Perny, M.; Ting, C.-C.; Kleinlogel, S.; Senn, P.; Roccio, M. Generation of Otic Sensory Neurons from Mouse Embryonic Stem Cells in 3D Culture. Front. Cell. Neurosci. 2017, 11, 409. [Google Scholar] [CrossRef] [Green Version]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercurio, S.; Serra, L.; Nicolis, S.K. More than just Stem Cells: Functional Roles of the Transcription Factor Sox2 in Differentiated Glia and Neurons. Int. J. Mol. Sci. 2019, 20, 4540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusznak, Z.; Szucs, G. Spiral ganglion neurones: An overview of morphology, firing behaviour, ionic channels and function. Pflug. Arch. 2009, 457, 1303–1325. [Google Scholar] [CrossRef]
- Mo, L.Z.; Davis, R.L. Endogenous firing patterns of murine spiral ganglion neurons. J. Neurophysiol. 1997, 77, 1294–1305. [Google Scholar] [CrossRef] [Green Version]
- Reid, M.A.; Flores-Otero, J.; Davis, R.L. Firing patterns of type II spiral ganglion neurons in vitro. J. Neurosci. 2004, 24, 733–742. [Google Scholar] [CrossRef] [Green Version]
- Adamson, C.L.; Reid, M.A.; Mo, Z.-L.; Bowne-English, J.; Davis, R.L. Firing features and potassium channel content of murine spiral ganglion neurons vary with cochlear location. J. Comp. Neurol. 2002, 447, 331–350. [Google Scholar] [CrossRef]
- Jagger, J.D.; Housley, G.D. Membrane properties of type II spiral ganglion neurones identified in a neonatal rat cochlear slice. J. Physiol. 2003, 552, 525–533. [Google Scholar] [CrossRef]
- Matsuoka, A.J.; Morrissey, Z.D.; Zhang, C.; Homma, K.; Belmadani, A.; Miller, C.A.; Chadly, D.M.; Kobayashi, S.; Edelbrock, A.N.; Tanaka-Matakatsu, M.; et al. Directed Differentiation of Human Embryonic Stem Cells Toward Placode-Derived Spiral Ganglion-Like Sensory Neurons. STEM CELLS Transl. Med. 2016, 6, 923–936. [Google Scholar] [CrossRef]
- Wang, M.; Xu, L.; Han, Y.; Wang, X.; Chen, F.; Lu, J.; Wang, H.; Liu, W. Regulation of Spiral Ganglion Neuron Regeneration as a Therapeutic Strategy in Sensorineural Hearing Loss. Front. Mol. Neurosci. 2022, 14, 829564. [Google Scholar] [CrossRef]
- Chang, S.-Y.; Jeong, H.-W.; Kim, E.; Jung, J.Y.; Lee, M.Y. Distribution and Afferent Effects of Transplanted mESCs on Cochlea in Acute and Chronic Neural Hearing Loss Models. BioMed Res. Int. 2021, 2021, 4956404. [Google Scholar] [CrossRef]
- Chen, W.; Jongkamonwiwat, N.; Abbas, L.; Eshtan, S.J.; Johnson, S.L.; Kuhn, S.; Milo, M.; Thurlow, J.K.; Andrews, P.W.; Marcotti, W.; et al. Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature 2012, 490, 278–282. [Google Scholar] [CrossRef]
- Li, X.; Bi, Z.; Sun, Y.; Li, C.; Li, Y.; Liu, Z. In vivo ectopic Ngn1 and Neurod1 convert neonatal cochlear glial cells into spiral ganglion neurons. FASEB J. 2020, 34, 4764–4782. [Google Scholar] [CrossRef]
- Kempfle, J.S.; Luu, N.-N.C.; Petrillo, M.; Al-Asad, R.; Zhang, A.; Edge, A.S.B. Lin28 reprograms inner ear glia to a neuronal fate. Stem Cells 2020, 38, 890–903. [Google Scholar] [CrossRef]
- Noda, T.; Meas, S.; Nogami, J.; Amemiya, Y.; Uchi, R.; Ohkawa, Y.; Nishimura, K.; Dabdoub, A. Direct Reprogramming of Spiral Ganglion Non-neuronal Cells into Neurons: Toward Ameliorating Sensorineural Hearing Loss by Gene Therapy. Front. Cell Dev. Biol. 2018, 6, 16. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Huang, Y.; Yu, C.; Liu, Q.; Qiu, C.; Wan, G. Cochlear Sox2+ Glial Cells Are Potent Progenitors for Spiral Ganglion Neuron Reprogramming Induced by Small Molecules. Front. Cell Dev. Biol. 2021, 9, 728352. [Google Scholar] [CrossRef]
- Samarajeewa, A.; Lenz, D.R.; Xie, L.; Chiang, H.; Kirchner, R.; Mulvaney, J.F.; Edge, A.S.B.; Dabdoub, A. Transcriptional response to Wnt activation regulates the regenerative capacity of the mammalian cochlea. Development 2018, 145, dev166579. [Google Scholar] [CrossRef] [Green Version]
- McLean, W.J.; Yin, X.; Lu, L.; Lenz, D.R.; McLean, D.; Langer, R.; Karp, J.M.; Edge, A.S. Clonal Expansion of Lgr5-Positive Cells from Mammalian Cochlea and High-Purity Generation of Sensory Hair Cells. Cell Rep. 2017, 18, 1917–1929. [Google Scholar] [CrossRef] [Green Version]
- Geng, R.; Noda, T.; Mulvaney, J.; Lin, V.Y.W.; Edge, A.S.B.; Dabdoub, A. Comprehensive Expression of Wnt Signaling Pathway Genes during Development and Maturation of the Mouse Cochlea. PLoS ONE 2016, 11, e0148339. [Google Scholar] [CrossRef] [Green Version]
- Jen, H.-I.; Hill, M.; Tao, L.; Sheng, K.; Cao, W.; Zhang, H.; Yu, H.; Llamas, J.; Zong, C.; Martin, J.F.; et al. Transcriptomic and epigenetic regulation of hair cell regeneration in the mouse utricle and its potentiation by Atoh1. eLife 2019, 8, e44328. [Google Scholar] [CrossRef]
- Kaiser, O.; Paasche, G.; Stöver, T.; Ernst, S.; Lenarz, T.; Kral, A.; Warnecke, A. TGF-beta superfamily member activin A acts with BDNF and erythropoietin to improve survival of spiral ganglion neurons in vitro. Neuropharmacology 2013, 75, 416–425. [Google Scholar] [CrossRef]
- Marzella, P.L.; Gillespie, L.N.; Clark, G.M.; Bartlett, P.F.; Kilpatrick, T.J. The neurotrophins act synergistically with LIF and members of the TGF-beta superfamily to promote the survival of spiral ganglia neurons in vitro. Hear. Res. 1999, 138, 73–80. [Google Scholar] [CrossRef]
- Marzella, P.L.; Clark, G.M.; Shepherd, R.K.; Bartlett, P.F.; Kilpatrick, T.J. Synergy between TGF-β3 and NT-3 to promote the survival of spiral ganglia neurones in vitro. Neurosci. Lett. 1998, 240, 77–80. [Google Scholar] [CrossRef]
- Meyers, A.E.; Kessler, J.A. TGF-β Family Signaling in Neural and Neuronal Differentiation, Development, and Function. Cold Spring Harb. Perspect. Biol. 2017, 9, a022244. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Zhang, H.; Yung, A.; Villeda, S.A.; Jaeger, P.A.; Olayiwola, O. ALK5-dependent TGF-β signaling is a major determinant of late-stage adult neurogenesis. Nat. Neurosci. 2014, 17, 943–952. [Google Scholar] [CrossRef] [Green Version]
- Aigner, L.; Wachs, F.P.; Winner, B.; Couillard-Despres, S.; Schiller, T.; Aigner, R.; Winkler, J.; Bogdahn, U. Transforming growth factor-beta1 is a negative modulator of adult neurogenesis. J. Neuropathol. Exp. Neurol. 2006, 65, 358–370. [Google Scholar]
- Liu, Q.; Zhang, L.; Zhu, M.-S.; Wan, G. High-throughput screening on cochlear organoids identifies VEGFR-MEK-TGFB1 signaling promoting hair cell reprogramming. Stem Cell Rep. 2021, 16, 2257–2273. [Google Scholar] [CrossRef] [PubMed]
- Koehler, K.R.; Nie, J.; Longworth-Mills, E.; Liu, X.-P.; Lee, J.; Holt, J.R.; Hashino, E. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 2017, 35, 583–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roccio, M. Directed differentiation and direct reprogramming: Applying stem cell technologies to hearing research. Stem Cells 2021, 39, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Roccio, M.; Edge, A.S.B. Inner ear organoids: New tools to understand neurosensory cell development, degeneration and regeneration. Development 2019, 146, dev177188. [Google Scholar] [CrossRef] [Green Version]
- Czajkowski, A.; Mounier, A.; Delacroix, L.; Malgrange, B. Pluripotent stem cell-derived cochlear cells: A challenge in constant progress. Experientia 2018, 76, 627–635. [Google Scholar] [CrossRef] [Green Version]
- Kurihara, S.; Fujioka, M.; Hirabayashi, M.; Yoshida, T.; Hosoya, M.; Nagase, M.; Kato, F.; Ogawa, K.; Okano, H.; Kojima, H.; et al. Otic Organoids Containing Spiral Ganglion Neuron-like Cells Derived from Human-induced Pluripotent Stem Cells as a Model of Drug-induced Neuropathy. Stem Cells Transl. Med. 2022, 11, 282–296. [Google Scholar] [CrossRef]
- Tang, P.-C.; Alex, A.L.; Nie, J.; Lee, J.; Roth, A.A.; Booth, K.T.; Koehler, K.R.; Hashino, E.; Nelson, R.F. Defective Tmprss3-Associated Hair Cell Degeneration in Inner Ear Organoids. Stem Cell Rep. 2019, 13, 147–162. [Google Scholar] [CrossRef] [Green Version]
- Stojkovic, M.; Han, D.; Jeong, M.; Stojkovic, P.; Stankovic, K.M. Human induced pluripotent stem cells and CRISPR/Cas-mediated targeted genome editing: Platforms to tackle sensorineural hearing loss. Stem Cells 2021, 39, 673–696. [Google Scholar] [CrossRef]







| Cell Type | Phoenix | si-ANs | Primary SGN | Literature 1°SGN | ||
|---|---|---|---|---|---|---|
| Type 1 | Type 2 | Type 1 | Type 1 | Type 2 | ||
| Abundance | 91% | 9% | 100% | 90–95% | 5–10% | [28] |
| Vm (mV) | −40.9 ± 1.5 | −43.3 ± 1.5 | −40.2 ± 1.9 | −60.5 ± 0.9 | −65.3 ± 1.7 | [30] |
| R (input) (MΩ) | 438 ± 39 | 233 ± 22 | 496 ± 87 | apex ~470 300 MΩ 510 ± 70 | Basal ~280 200 MΩ 360 ± 120 | [31] [28] [32] |
| Spike latency (ms) | 3.1 ± 0.4 | 2.1 ± 0.1 | 4.4 ± 0.8 | 4.2 ± 0.8 | 4.8 ± 0.5 | [32] |
| AP amplitude (mV) | 34.3 ± 10.7 | 49.2 ± 11.6 | 29.0 ± 5.4 | 50–95 | [33] | |
| AP width (ms) | 2.2 ± 0.2 | 1.3 ± 0.2 | 2.5 ± 0.1 | Apex: 1.9 ± 0.01 Base: 1.68 ± 0.2 | Apex: 1.4 ± 0.02 Base: 1.25 ± 0.03 | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rousset, F.; Schilardi, G.; Sgroi, S.; Nacher-Soler, G.; Sipione, R.; Kleinlogel, S.; Senn, P. WNT Activation and TGFβ-Smad Inhibition Potentiate Stemness of Mammalian Auditory Neuroprogenitors for High-Throughput Generation of Functional Auditory Neurons In Vitro. Cells 2022, 11, 2431. https://doi.org/10.3390/cells11152431
Rousset F, Schilardi G, Sgroi S, Nacher-Soler G, Sipione R, Kleinlogel S, Senn P. WNT Activation and TGFβ-Smad Inhibition Potentiate Stemness of Mammalian Auditory Neuroprogenitors for High-Throughput Generation of Functional Auditory Neurons In Vitro. Cells. 2022; 11(15):2431. https://doi.org/10.3390/cells11152431
Chicago/Turabian StyleRousset, Francis, Giulia Schilardi, Stéphanie Sgroi, German Nacher-Soler, Rebecca Sipione, Sonja Kleinlogel, and Pascal Senn. 2022. "WNT Activation and TGFβ-Smad Inhibition Potentiate Stemness of Mammalian Auditory Neuroprogenitors for High-Throughput Generation of Functional Auditory Neurons In Vitro" Cells 11, no. 15: 2431. https://doi.org/10.3390/cells11152431