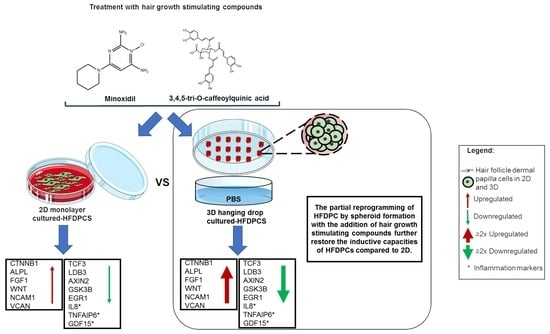
3D Spheroid Human Dermal Papilla Cell as an Effective Model for the Screening of Hair Growth Promoting Compounds: Examples of Minoxidil and 3,4,5-Tri-O-caffeoylquinic acid (TCQA)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Preparation
2.2. Cells and Cell Culture
2.3. Establishment and Maintenance of 3D Culture
2.4. RNA Extraction
2.5. DNA Microarray Analysis
2.6. Quantitative Real-Time PCR Analysis
2.7. Western Blot
2.8. Statistical Analysis
3. Results
3.1. Gene Profiling Analysis of HFDPCs Cultured in 3D Hanging Drops Compared with 2D Culture
3.2. Comparison of the DEGs in HFDPCs Treated with Minoxidil and TCQA in Both Systems
3.3. Comparison of DEG between 3D and 2D System upon Minoxidil Treatment
3.4. 3D Cultured DPCs Treated with Minoxidil Enhanced the Gene and Protein Expression of Hair Growth Markers
3.5. Characteristics of 2D and 3D Cultured HFDPCs Treated with TCQA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schneider, M.R.; Schmidt-Ullrich, R.; Paus, R. The Hair Follicle as a Dynamic Miniorgan. Curr. Biol. 2009, 19, R132–R142. [Google Scholar] [CrossRef] [Green Version]
- Stenn, K.S.; Paus, R. Controls of hair follicle cycling. Physiol. Rev. 2001, 81, 449–494. [Google Scholar] [CrossRef]
- Paus, R.; Cotsarelis, G. The Biology of Hair Follicles. N. Engl. J. Med. 1999, 341, 491–497. [Google Scholar] [CrossRef] [Green Version]
- Malkud, S. Telogen Effluvium: A Review. J. Clin. Diagn. Res. 2015, 9, WE01–WE03. [Google Scholar] [CrossRef]
- Enshell-Seijffers, D.; Lindon, C.; Kashiwagi, M.; Morgan, B.A. β-catenin Activity in the Dermal Papilla Regulates Morphogenesis and Regeneration of Hair. Dev. Cell 2010, 18, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Truong, V.-L.; Bak, M.J.; Lee, C.; Jun, M.; Jeong, W.-S. Hair Regenerative Mechanisms of Red Ginseng Oil and Its Major Components in the Testosterone-Induced Delay of Anagen Entry in C57BL/6 Mice. Molecules 2017, 22, 1505. [Google Scholar] [CrossRef] [Green Version]
- Driskell, R.R.; Clavel, C.; Rendl, M.; Watt, F.M. Hair follicle dermal papilla cells at a glance. J. Cell Sci. 2011, 124, 1179–1182. [Google Scholar] [CrossRef] [Green Version]
- Betriu, N.; Jarrosson-Moral, C.; Semino, C.E. Culture and Differentiation of Human Hair Follicle Dermal Papilla Cells in a Soft 3D Self-Assembling Peptide Scaffold. Biomolecules 2020, 10, 684. [Google Scholar] [CrossRef]
- Higgins, C.A.; Chen, J.C.; Cerise, J.E.; Jahoda, C.A.B.; Christiano, A.M. Microenvironmental reprogramming by threedimensional culture enables dermal papilla cells to induce de novo human hair-follicle growth. Proc. Natl. Acad. Sci. USA 2013, 110, 19679–19688. [Google Scholar] [CrossRef] [Green Version]
- Botchkarev, V.A.; Kishimoto, J. Molecular control of epithelial-mesenchymal interactions during hair follicle cycling. In Proceedings of the Journal of Investigative Dermatology Symposium Proceedings; Elsevier Masson SAS: Ile De France, France, 2003; Volume 8, pp. 46–55. [Google Scholar]
- Weaver, V.M.; Petersen, O.W.; Wang, F.; Larabell, C.A.; Briand, P.; Damsky, C.; Bissell, M.J. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 1997, 137, 231–245. [Google Scholar] [CrossRef] [Green Version]
- Bhadriraju, K.; Chen, C.S. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discov. Today 2002, 7, 612–620. [Google Scholar] [CrossRef]
- Baharvand, H.; Hashemi, S.M.; Kazemi Ashtiani, S.; Farrokhi, A. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int. J. Dev. Biol. 2006, 50, 645–652. [Google Scholar] [CrossRef] [Green Version]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Benya, P.D.; Shaffer, J.D. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell 1982, 30, 215–224. [Google Scholar] [CrossRef]
- Nelson, C.M.; Bissell, M.J. Modeling dynamic reciprocity: Engineering three-dimensional culture models of breast architecture, function, and neoplastic transformation. Semin. Cancer Biol. 2005, 15, 342–352. [Google Scholar] [CrossRef] [Green Version]
- Shield, K.; Ackland, M.L.; Ahmed, N.; Rice, G.E. Multicellular spheroids in ovarian cancer metastases: Biology and pathology. Gynecol. Oncol. 2009, 113, 143–148. [Google Scholar] [CrossRef]
- Zietarska, M.; Maugard, C.M.; Filali-Mouhim, A.; Alam-Fahmy, M.; Tonin, P.N.; Provencher, D.M.; Mes-Masson, A.-M. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC). Mol. Carcinog. 2007, 46, 872–885. [Google Scholar] [CrossRef]
- Lin, B.; Miao, Y.; Wang, J.; Fan, Z.; Du, L.; Su, Y.; Liu, B.; Hu, Z.; Xing, M. Surface Tension Guided Hanging-Drop: Producing Controllable 3D Spheroid of High-Passaged Human Dermal Papilla Cells and Forming Inductive Microtissues for Hair-Follicle Regeneration. ACS Appl. Mater. Interfaces 2016, 8, 5906–5916. [Google Scholar] [CrossRef]
- Ohyama, M.; Zheng, Y.; Paus, R.; Stenn, K.S. The mesenchymal component of hair follicle neogenesis: Background, methods and molecular characterization. Exp. Dermatol. 2010, 19, 89–99. [Google Scholar] [CrossRef]
- Lodi, G.; Sannino, M.; Cannarozzo, G.; Giudice, A.; Del Duca, E.; Tamburi, F.; Bennardo, L.; Nisticò, S.P. Blue light-emitting diodes in hair regrowth: The first prospective study. Lasers Med. Sci. 2021, 36, 1719–1723. [Google Scholar] [CrossRef]
- Nistico, S.; Tamburi, F.; Bennardo, L.; Dastoli, S.; Schipani, G.; Caro, G.; Fortuna, M.C.; Rossi, A. Treatment of telogen effluvium using a dietary supplement containing Boswellia serrata, Curcuma longa, and Vitis vinifera: Results of an observational study. Dermatol. Ther. 2019, 32, e12842. [Google Scholar] [CrossRef]
- Bejaoui, M.; Villareal, M.O.; Isoda, H. 3,4,5-Tri-O-Caffeoylquinic Acid Promoted Hair Pigmentation Through β-Catenin and Its Target Genes. Front. Cell Dev. Biol. 2020, 8, 175. [Google Scholar] [CrossRef] [PubMed]
- Bejaoui, M.; Villareal, M.O.; Isoda, H. β-catenin-mediated hair growth induction effect of 3,4,5-tri-O-caffeoylquinic acid. Aging 2019, 11, 4216–4237. [Google Scholar] [CrossRef] [PubMed]
- Bejaoui, M.; Ferdousi, F.; Zheng, Y.-W.; Oda, T.; Isoda, H. Regulating cell fate of human amnion epithelial cells using natural compounds: An example of enhanced neural and pigment differentiation by 3,4,5-tri-O-caffeoylquinic acid. Cell Commun. Signal. 2021, 19, 26. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Li, Z.; Lutz, H.; Huang, K.; Su, T.; Cores, J.; Dinh, P.-U.C.; Cheng, K. Dermal exosomes containing miR-218-5p promote hair regeneration by regulating β-catenin signaling. Sci. Adv. 2020, 6, eaba1685. [Google Scholar] [CrossRef]
- Jahoda, C.A.B.; Horne, K.A.; Oliver, R.F. Induction of hair growth by implantation of cultured dermal papilla cells. Nature 1984, 311, 560–562. [Google Scholar] [CrossRef]
- Jahoda, C.A.B.; Oliver, R.F. Vibrissa dermal papilla cell aggregative behaviour in vivo and in vitro. J. Embryol. Exp. Morphol. 1984, 79, 211–224. [Google Scholar] [CrossRef]
- Schmidt-Ullrich, R.; Paus, R. Molecular principles of hair follicle induction and morphogenesis. BioEssays 2005, 27, 247–261. [Google Scholar] [CrossRef]
- Osada, A.; Iwabuchi, T.; Kishimoto, J.; Hamazaki, T.S.; Okochi, H. Long-Term Culture of Mouse Vibrissal Dermal Papilla Cells and De Novo Hair Follicle Induction. Tissue Eng. 2007, 13, 975–982. [Google Scholar] [CrossRef]
- Jahoda, C.A.B. Cellular and developmental aspects of androgenetic alopecia. Exp. Dermatol. 1998, 7, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Messenger, A.G.; Jennifer, H.; Bleehen, S.S. The in vitro properties of dermal papilla cell lines established from human hair follicles. Br. J. Dermatol. 1986, 114, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Young, T.-H.; Tu, H.-R.; Chan, C.-C.; Huang, Y.-C.; Yen, M.-H.; Cheng, N.-C.; Chiu, H.-C.; Lin, S.-J. The enhancement of dermal papilla cell aggregation by extracellular matrix proteins through effects on cell-substratum adhesivity and cell motility. Biomaterials 2009, 30, 5031–5040. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Sun, Y.B.; Liu, B.C.; Jiang, J.D.; Hu, Z.Q. Controllable production of transplantable adult human high-passage dermal papilla spheroids using 3D matrigel culture. Tissue Eng. Part A 2014, 20, 2329–2338. [Google Scholar] [CrossRef] [Green Version]
- Rendl, M.; Lewis, L.; Fuchs, E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005, 3, e331. [Google Scholar] [CrossRef]
- Madaan, A.; Verma, R.; Singh, A.T.; Jaggi, M. Review of Hair Follicle Dermal Papilla cells as in vitro screening model for hair growth. Int. J. Cosmet. Sci. 2018, 40, 429–450. [Google Scholar] [CrossRef] [Green Version]
- Combates, N.J.; Chuong, C.-M.; Stenn, K.S.; Prouty, S.M. Expression of Two Ig Family Adhesion Molecules in the Murine Hair Cycle: DCC in the Bulge Epithelia and NCAM in the Follicular Papilla. J. Investig. Dermatol. 1997, 109, 672–678. [Google Scholar] [CrossRef] [Green Version]
- Ting-Berreth, S.A.; Chuong, C.-M. Sonic hedgehog in feather morphogenesis: Induction of mesenchymal condensation and association with cell death. Dev. Dyn. 1996, 207, 157–170. [Google Scholar] [CrossRef]
- Du Cros, D.L.; LeBaron, R.G.; Couchman, J.R. Association of versican with dermal matrices and its potential role in hair follicle development and cycling. J. Investig. Dermatol. 1995, 105, 426–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, N.; Tanigaki, K.; Han, H.; Hiai, H.; Honjo, T. Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr. Biol. 2003, 13, 333–338. [Google Scholar] [CrossRef] [Green Version]
- Blanpain, C.; Lowry, W.E.; Pasolli, H.A.; Fuchs, E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006, 20, 3022–3035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philpott, M.P.; Sanders, D.A.; Bowen, J.; Kealey, T. Effects of interleukins, colony-stimulating factor and tumour necrosis factor on human hair follicle growth in vitro: A possible role for interleukin-1 and tumour necrosis factor-alpha in alopecia areata. Br. J. Dermatol. 1996, 135, 942–948. [Google Scholar] [CrossRef]
- Kwack, M.H.; Ahn, J.S.; Kim, M.K.; Kim, J.C.; Sung, Y.K. Dihydrotestosterone-inducible IL-6 inhibits elongation of human hair shafts by suppressing matrix cell proliferation and promotes regression of hair follicles in mice. J. Investig. Dermatol. 2012, 132, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Higgins, C.A.; Richardson, G.D.; Ferdinando, D.; Westgate, G.E.; Jahoda, C.A.B. Modelling the hair follicle dermal papilla using spheroid cell cultures. Exp. Dermatol. 2010, 19, 546–548. [Google Scholar] [CrossRef]
- Griffith, L.G.; Swartz, M.A. Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 2006, 7, 211–224. [Google Scholar] [CrossRef]
- Campuzano, S.; Pelling, A.E. Scaffolds for 3D Cell Culture and Cellular Agriculture Applications Derived from Non-animal Sources. Front. Sustain. Food Syst. 2019, 3, 38. [Google Scholar] [CrossRef] [Green Version]
- Andersen, T.; Auk-Emblem, P.; Dornish, M. 3D Cell Culture in Alginate Hydrogels. Microarrays 2015, 4, 133–161. [Google Scholar] [CrossRef]
- Mastrorocco, A.; Cacopardo, L.; Lamanna, D.; Temerario, L.; Brunetti, G.; Carluccio, A.; Robbe, D.; Dell’Aquila, M.E. Bioengineering Approaches to Improve In Vitro Performance of Prepubertal Lamb Oocytes. Cells 2021, 10, 1458. [Google Scholar] [CrossRef]
- Yamauchi, K.; Kurosaka, A. Inhibition of glycogen synthase kinase-3 enhances the expression of alkaline phosphatase and insulin-like growth factor-1 in human primary dermal papilla cell culture and maintains mouse hair bulbs in organ culture. Arch. Dermatol. Res. 2009, 301, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Abreu, C.M.; Cerqueira, M.T.; Pirraco, R.P.; Gasperini, L.; Reis, R.L.; Marques, A.P. Rescuing key native traits in cultured dermal papilla cells for human hair regeneration. J. Adv. Res. 2021, 30, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Mori, O.; Uno, H. The effect of topical minoxidil on hair follicular cycles of rats. J. Dermatol. 1990, 17, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Price, V.H.; Menefee, E.; Strauss, P.C. Changes in hair weight and hair count in men with androgenetic alopecia, after application of 5% and 2% topical minoxidil, placebo, or no treatment. J. Am. Acad. Dermatol. 1999, 41, 717–721. [Google Scholar] [CrossRef]
- Suchonwanit, P.; Thammarucha, S.; Leerunyakul, K. Minoxidil and its use in hair disorders: A review. Drug Des. Devel. Ther. 2019, 13, 2777–2786. [Google Scholar] [CrossRef] [Green Version]
- Messenger, A.G.; Rundegren, J. Minoxidil: Mechanisms of action on hair growth. Br. J. Dermatol. 2004, 150, 186–194. [Google Scholar] [CrossRef]
- Shin, H.; Kwack, M.H.; Shin, S.H.; Oh, J.W.; Kang, B.M.; Kim, A.A.; Kim, J.; Kim, M.K.; Kim, J.C.; Sung, Y.K. Identification of transcriptional targets of Wnt/beta-catenin signaling in dermal papilla cells of human scalp hair follicles: EP2 is a novel transcriptional target of Wnt3a. J. Dermatol. Sci. 2010, 58, 91–96. [Google Scholar] [CrossRef]
- Rajendran, R.L.; Gangadaran, P.; Bak, S.S.; Oh, J.M.; Kalimuthu, S.; Lee, H.W.; Baek, S.H.; Zhu, L.; Sung, Y.K.; Jeong, S.Y.; et al. Extracellular vesicles derived from MSCs activates dermal papilla cell in vitro and promotes hair follicle conversion from telogen to anagen in mice. Sci. Rep. 2017, 7, 15560. [Google Scholar] [CrossRef] [Green Version]
- Adachi, K.; Watanabe, Y.; Inouye, K. Activity of glucose-6-phosphate 1-dehydrogenase in hair follicles with male-pattern alopecia. Biosci. Biotechnol. Biochem. 1999, 63, 2219–2221. [Google Scholar] [CrossRef] [Green Version]
- Ohyama, M.; Kobayashi, T.; Sasaki, T.; Shimizu, A.; Amagai, M. Restoration of the intrinsic properties of human dermal papilla in vitro. J. Cell Sci. 2012, 125, 4114–4125. [Google Scholar] [CrossRef] [Green Version]
- Soma, T.; Tajima, M.; Kishimoto, J. Hair cycle-specific expression of versican in human hair follicles. J. Dermatol. Sci. 2005, 39, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Cha, S.Y.; Kim, M.K.; Kim, J.C.; Sung, Y.K. Induction of versican by ascorbic acid 2-phosphate in dermal papilla cells. J. Dermatol. Sci. 2006, 43, 60–62. [Google Scholar] [CrossRef] [PubMed]
- Takabayashi, Y.; Nambu, M.; Ishihara, M.; Kuwabara, M.; Fukuda, K.; Nakamura, S.; Hattori, H.; Kiyosawa, T. Enhanced effect of fibroblast growth factor-2-containing dalteparin/protamine nanoparticles on hair growth. Clin. Cosmet. Investig. Dermatol. 2016, 9, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Xiang, L.-J.; Shi, H.-X.; Zhang, J.; Jiang, L.; Cai, P.; Lin, Z.-L.; Lin, B.-B.; Huang, Y.; Zhang, H.-L.; et al. Fibroblast Growth Factors Stimulate Hair Growth throughβ-Catenin and Shh Expression in C57BL/6 Mice. Biomed Res. Int. 2015, 2015, 730139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maretto, S.; Cordenonsi, M.; Dupont, S.; Braghetta, P.; Broccoli, V.; Hassan, A.B.; Volpin, D.; Bressan, G.M.; Piccolo, S. Mapping Wnt/ -catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 3299–3304. [Google Scholar] [CrossRef] [Green Version]
- Andl, T.; Reddy, S.T.; Gaddapara, T.; Millar, S.E. WNT signals are required for the initiation of hair follicle development. Dev. Cell 2002, 2, 643–653. [Google Scholar] [CrossRef]
- Topouzi, H.; Logan, N.J.; Williams, G.; Higgins, C.A. Methods for the isolation and 3D culture of dermal papilla cells from human hair follicles. Exp. Dermatol. 2017, 26, 491–496. [Google Scholar] [CrossRef]
- Hu, S.; Li, Z.; Lutz, H.; Huang, K.; Su, T.; Cores, J.; Dinh, P.-U.C.; Cheng, K.; Botchkarev, V.A.; Kishimoto, J.; et al. Culture and Differentiation of Human Hair Follicle Dermal Papilla Cells in a Soft 3D Self-Assembling Peptide Scaffold. J. Dermatol. Sci. 2010, 19, 672–678. [Google Scholar]
- Horsley, V.; Aliprantis, A.O.; Polak, L.; Glimcher, L.H.; Fuchs, E. NFATc1 Balances Quiescence and Proliferation of Skin Stem Cells. Cell 2008, 132, 299–310. [Google Scholar] [CrossRef] [Green Version]
- Keyes, B.E.; Segal, J.P.; Heller, E.; Lien, W.-H.; Chang, C.-Y.; Guo, X.; Oristian, D.S.; Zheng, D.; Fuchs, E. Nfatc1 orchestrates aging in hair follicle stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, E4950–E4959. [Google Scholar] [CrossRef] [Green Version]
- Avigad Laron, E.; Aamar, E.; Enshell-Seijffers, D. The Mesenchymal Niche of the Hair Follicle Induces Regeneration by Releasing Primed Progenitors from Inhibitory Effects of Quiescent Stem Cells. Cell Rep. 2018, 24, 909–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, B.Y. Targeting Wnt/β-Catenin Pathway for Developing Therapies for Hair Loss. Int. J. Mol. Sci. 2020, 21, 4915. [Google Scholar] [CrossRef] [PubMed]
- Katsuoka, K.; Mauch, C.; Schell, H.; Hornstein, O.P.; Krieg, T. Collagen-type synthesis in human-hair papilla cells in culture. Arch. Dermatol. Res. 1988, 280, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Tsang, K.Y.; Cheung, M.C.H.; Chan, D.; Cheah, K.S.E. The developmental roles of the extracellular matrix: Beyond structure to regulation. Cell Tissue Res. 2009, 339, 93–110. [Google Scholar] [CrossRef] [Green Version]
- Malgouries, S.; Thibaut, S.; Bernard, B.A. Proteoglycan expression patterns in human hair follicle. Br. J. Dermatol. 2008, 158, 234–342. [Google Scholar] [CrossRef]





| Gene Symbol | Gene Name | Biological Function | Fold-Change | p Value ** |
|---|---|---|---|---|
| OXTR | Oxytocin receptor | Cell-surface-receptor signalling pathway; oxidative stress | 24.82 | 0.0002 |
| COL11A1 | Collagen alpha-1 (XI) chain | Extracellular matrix binding | 21.67 | 0.003 |
| SSTR1 | Somatostatin receptor type 1 | Expressed during anagen; HF immune system | 21.03 | 0.001 |
| NEDD9 | Enhancer of filamentation 1 | Cell adhesion; hair placode signature genes | 18.48 | 0.004 |
| HAPLN1 | Hyaluronan and proteoglycan link protein 1 | Cell adhesion; cell–cell communication | 14.06 | 0.005 |
| IL8 | Interleukin-8 | Immune response; inflammation | −179.99 | 0.004 |
| TNFAIP6 | Tumor necrosis factor-inducible gene 6 protein | TNF-α signalling; inflammation | −32 | 0.001 |
| RANBP3L | Ran-binding protein 3-like | Termination of BMP signalling; inhibition of mesenchymal stem cell differentiation | −31.18 | 0.003 |
| GDF15 | Growth/differentiation factor 15 | Stress response cytokine, expression increases with inflammation and injury | −25.96 | 0.045 |
| Gene Symbol | Gene Name | Biological Function | Fold-Change | p Value ** |
|---|---|---|---|---|
| CTNNB1 | Catenin (cadherin associated protein), beta 1 | Anagen regulation; hair follicle morphogenesis; positive regulation of fibroblast growth; dermal papilla cell proliferation | 19.06 | 0.049 |
| TRH | Pro-thyrotropin-releasing hormone | Hair shaft elongation; anagen prolongation; hair matrix keratinocyte proliferation | 7.25 | 0.011 |
| RASGRF1 | Ras-specific guanine nucleotide-releasing factor | Hair follicle morphogenesis; ATP binding | 6.75 | 0.020 |
| FERMT1 | Fermitin family homolog 1 | Cell adhesion; keratinocyte proliferation and morphogenesis | 6.03 | 0.018 |
| ABCC11 | ATP-binding cassette sub-family C member 11 | ATP binding | 5.88 | 0.029 |
| CXXC5 | CXXC-type zinc finger protein 5 | Involved in MAPK pathway; cell cycle arrest | 5.11 | 0.021 |
| NCAM | Neural cell adhesion molecule | Hair morphogenesis; highly expressed during anagen; keratinocyte segregation and differentiation | 4.72 | 0.085 |
| COL28A1 | Collagen alpha-1(XXVIII) chain | Hair shaft strength; matrix assembly | 4.62 | 0.006 |
| KRT40 | Keratin, type I cytoskeletal 40 | Late hair differentiation | 4.06 | 0.010 |
| VCAN | Versican | Collagen-containing extracellular matrix; highly expressed during anagen | 3.23 | 0.008 |
| Gene Symbol | Gene Name | Biological Function | Fold-Change | p Value ** |
|---|---|---|---|---|
| NFATC1 | Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 | Stem cell quiescence | −8.22 | 0.001 |
| STEAP3 | STEAP family member 3 | Inhibition of cell cycle arrest; downregulation of P53 pathway | −7.96 | 0.003 |
| LFNG | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | Notch pathway inhibition; decreased hair development | −7.07 | 0.011 |
| PRUNE2 | Protein prune homolog 2 | Regulation of tumour cell differentiation, survival, and aggressiveness | −6.82 | 0.032 |
| SKIV21 | Superkiller viralicidic activity 2-lik | ATP degradation | −5.45 | 0.007 |
| TRABD2B | TraB domain containing 2B | Negative regulator of the Wnt pathway; Wnt protein cleavage | −5.04 | 0.027 |
| MAF | Avian musculoaponeurotic fibrosarcoma oncogene homolog | Oncogene; embryonic marker of development | −4.78 | 0.062 |
| STYK1 | Serine/threonine/tyrosine kinase 1 | Tumour cell invasion and metastasis | −3.42 | 0.012 |
| AXIN2 | Axin 2 | β-Catenin phosphorylation and degradation | −3.33 | 0.028 |
| SIX3 | Sine oculis-related homeobox 3 | Wnt/β-Catenin repression | −2.74 | 0.005 |
| Gene Symbol | Gene Name | Biological Function | Fold-Change | ||
|---|---|---|---|---|---|
| 2D | 3D | In Vivo | |||
| CTNNB1 | Catenin (cadherin associated protein), beta 1 | Anagen regulation; hair follicle morphogenesis; positive regulation of fibroblast growth; dermal papilla cell proliferation | 1.62 | 43.57 | 3.2 |
| FGF1 | Fibroblast growth factor 1 | Hair follicle morphogenesis; hair growth cycle regulation | 1.53 | 26.45 | 3.22 |
| NRAS | Neuroblastoma RAS viral oncogene homolog | Cell proliferation | 1.53 | 16 | 1.2 |
| ALPL | Alkaline phosphatase | Wnt/β-catenin pathway regulator | 1.2 | 8 | 2.33 |
| BMPR-1A | Bone morphogenetic protein | Skin development; hair follicle growth | 1.14 | 64.89 | 1.7 |
| VCAN | Versican | Cell aggregation marker; cell adhesion and proliferation | 4.61 | 12.21 | 1.6 |
| WLS | Wntless | Wnt secretion and pathway | 1.6 | 16.38 | 3 |
| AXIN2 | Axis inhibition protein 2 | Wnt-responsive gene | −1.5 | −5 | −2.82 |
| TCF3 | Transcription factor 3 | Wnt/β-Catenin repression | −1.1 | −5 | −2.82 |
| LDB3 | LIM Domain Binding 3 | Wnt-responsive gene; regulation of hair follicle during telogen | −1.14 | −2.7 | −8.7 |
| EGR1 | Early growth response 1 | Negative regulation of Wnt/β-Catenin; upregulated with aging | −1.7 | −4 | −2.6 |
| GSK3B | Glycogen synthase kinase 3 beta | Phosphorylation of β-catenin | −1.5 | −6 | −5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bejaoui, M.; Oliva, A.K.; Ke, M.S.; Ferdousi, F.; Isoda, H. 3D Spheroid Human Dermal Papilla Cell as an Effective Model for the Screening of Hair Growth Promoting Compounds: Examples of Minoxidil and 3,4,5-Tri-O-caffeoylquinic acid (TCQA). Cells 2022, 11, 2093. https://doi.org/10.3390/cells11132093
Bejaoui M, Oliva AK, Ke MS, Ferdousi F, Isoda H. 3D Spheroid Human Dermal Papilla Cell as an Effective Model for the Screening of Hair Growth Promoting Compounds: Examples of Minoxidil and 3,4,5-Tri-O-caffeoylquinic acid (TCQA). Cells. 2022; 11(13):2093. https://doi.org/10.3390/cells11132093
Chicago/Turabian StyleBejaoui, Meriem, Aprill Kee Oliva, May Sin Ke, Farhana Ferdousi, and Hiroko Isoda. 2022. "3D Spheroid Human Dermal Papilla Cell as an Effective Model for the Screening of Hair Growth Promoting Compounds: Examples of Minoxidil and 3,4,5-Tri-O-caffeoylquinic acid (TCQA)" Cells 11, no. 13: 2093. https://doi.org/10.3390/cells11132093