Progression and Differentiation of Alveolar Rhabdomyosarcoma Is Regulated by PAX7 Transcription Factor—Significance of Tumor Subclones
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Transfection with siRNA
2.3. Proliferation, Cell Cycle, and BrdU Incorporation
2.4. Scratch Assay
2.5. Chemotaxis Assay
2.6. Flow Cytometry
2.7. RNA Isolation and Reverse Transcription
2.8. Quantitative Real-Time PCR
- PAX3-FOXO1 forward: 5′-AACCCCACCATTGGCAATG-3′
- PAX3-FOXO1 reverse: 5′-ACCCTCTGGATTGAGCATCCA-3′
2.9. Immunofluorescent Staining
2.10. In Vivo Experiments
2.11. Analysis of RMS Tumor Samples from Patients
2.12. Bioinformatic Analysis of Literature Data from RMS Cell Lines and Tumors
2.13. Bioinformatic Analysis of miRNA Targets
2.14. Statistical Analysis
3. Results
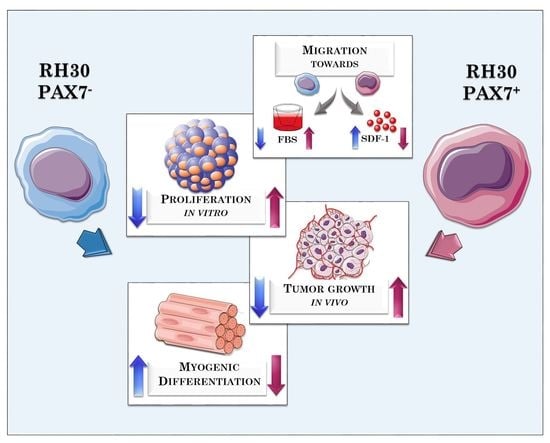
3.1. RH30 Subclones of Different Origin Display Differences in Morphology, Proliferation Rate, and Migration
3.2. RH30 Subclones Display Differences in Expression Levels of Surface Markers and Myogenic Transcription Factors
3.3. PAX7 Transcription Factor Regulates Rhabdomyosarcoma Proliferation, Migration, and Differentiation
3.4. ERMS and ARMS Cell Lines Differentially Express Transcription Factors and microRNA That May Be Associated with Myogenic Differentiation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, P.Y.; Guttridge, D.C. Dysregulated myogenesis in rhabdomyosarcoma. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 126, pp. 285–297. [Google Scholar]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef]
- Relaix, F.; Demignon, J.; Laclef, C.; Pujol, J.; Santolini, M.; Niro, C.; Lagha, M.; Rocancourt, D.; Buckingham, M.; Maire, P. Six Homeoproteins Directly Activate Myod Expression in the Gene Regulatory Networks That Control Early Myogenesis. PLoS Genet. 2013, 9, e1003425. [Google Scholar] [CrossRef] [Green Version]
- Grifone, R.; Demignon, J.; Houbron, C.; Souil, E.; Niro, C.; Seller, M.J.; Hamard, G.; Maire, P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 2005, 132, 2235–2249. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Chu, A.; Chakroun, I.; Islam, U.; Blais, A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res. 2010, 38, 6857–6871. [Google Scholar] [CrossRef] [Green Version]
- Ling, F.; Kang, B.; Sun, X.H. Id proteins: Small molecules, mighty regulators. In Current Topics in Developmental Biology; Academic Press Inc.: Cambridge, MA, USA, 2014; Volume 110, pp. 189–216. [Google Scholar]
- Langlands, K.; Yin, X.; Anand, G.; Prochownik, E.V. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J. Biol. Chem. 1997, 272, 19785–19793. [Google Scholar] [CrossRef] [Green Version]
- Schaaf, G.J.; Ruijter, J.M.; Ruissen, F.; Zwijnenburg, D.A.; Waaijer, R.; Valentijn, L.J.; Benit-Deekman, J.; Kampen, A.H.C.; Baas, F.; Kool, M. Full transcriptome analysis of rhabdomyosarcoma, normal and fetal skeletal muscle: Statistical comparison of multiple SAGE libraries. FASEB J. 2005, 19, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Prim. 2019, 5, 1. [Google Scholar] [CrossRef]
- Libura, J.; Drukala, J.; Majka, M.; Tomescu, O.; Navenot, J.M.; Kucia, M.; Marquez, L.; Peiper, S.C.; Barr, F.G.; Janowska-Wieczorek, A.; et al. CXCR4-SDF-1 signaling is active in rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and adhesion. Blood 2002, 100, 2597–2606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miekus, K.; Lukasiewicz, E.; Jarocha, D.; Sekula, M.; Drabik, G.; Majka, M. The decreased metastatic potential of rhabdomyosarcoma cells obtained through MET receptor downregulation and the induction of differentiation. Cell Death Dis. 2013, 4, e459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szewczyk, B.; Skrzypek, K.; Majka, M. Targeting MET Receptor in Rhabdomyosarcoma: Rationale and Progress. Curr. Drug Targets 2016, 18, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Skrzypek, K.; Kusienicka, A.; Szewczyk, B.; Adamus, T.; Lukasiewicz, E.; Miekus, K.; Majka, M. Constitutive activation of MET signaling impairs myogenic differentiation of rhabdomyosarcoma and promotes its development and progression. Oncotarget 2015, 6, 31378–31398. [Google Scholar] [CrossRef] [Green Version]
- Skrzypek, K.; Kot, M.; Konieczny, P.; Nieszporek, A.; Kusienicka, A.; Lasota, M.; Bobela, W.; Jankowska, U.; Kędracka-Krok, S.; Majka, M. SNAIL promotes metastatic behavior of rhabdomyosarcoma by increasing EZRIN and AKT expression and regulating microRNA networks. Cancers 2020, 12, 1870. [Google Scholar] [CrossRef]
- Skrzypek, K.; Kusienicka, A.; Trzyna, E.; Szewczyk, B.; Ulman, A.; Konieczny, P.; Adamus, T.; Badyra, B.; Kortylewski, M.; Majka, M. SNAIL is a key regulator of alveolar rhabdomyosarcoma tumor growth and differentiation through repression of MYF5 and MYOD function. Cell Death Dis. 2018, 9, 643. [Google Scholar] [CrossRef] [PubMed]
- Ulman, A.; Skrzypek, K.; Konieczny, P.; Mussolino, C.; Cathomen, T.; Majka, M. Genome Editing of the SNAI1 Gene in Rhabdomyosarcoma: A Novel Model for Studies of Its Role. Cells 2020, 9, 1095. [Google Scholar] [CrossRef]
- Cieśla, M.; Dulak, J.; Józkowicz, A. MicroRNAs and epigenetic mechanisms of rhabdomyosarcoma development. Int. J. Biochem. Cell Biol. 2014, 53C, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Skrzypek, K.; Majka, M. Interplay among SNAIL transcription factor, microRNAs, long non-coding RNAs, and circular RNAs in the regulation of tumor growth and metastasis. Cancers 2020, 12, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skrzypek, K.; Nieszporek, A.; Badyra, B.; Lasota, M.; Majka, M. Enhancement of myogenic differentiation and inhibition of rhabdomyosarcoma progression by miR-28-3p and miR-193a-5p regulated by SNAIL. Mol. Ther. Nucleic Acids 2021, 24, 888–904. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.A.; Garcia, M.R.; Go, J.C.; Finkelstein, D.; Kodali, K.; Pagala, V.; Wang, X.; Peng, J.; Hatley, M.E. PAX7 is a required target for microRNA-206-induced differentiation of fusion-negative rhabdomyosarcoma. Cell Death Dis. 2016, 7, e2256. [Google Scholar] [CrossRef]
- Gisselsson, D.; Lichtenzstejn, D.; Kachko, P.; Karlsson, J.; Manor, E.; Mai, S. Clonal evolution through genetic bottlenecks and telomere attrition: Potential threats to in vitro data reproducibility. Genes Chromosom. Cancer 2019, 58, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Porter, S.N.; Baker, L.C.; Mittelman, D.; Porteus, M.H. Lentiviral and targeted cellular barcoding reveals ongoing clonal dynamics of cell lines in vitro and in vivo. Genome Biol. 2014, 15, R75. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A.; Francisco, S. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Hou, W.; Epperly, M.W.; Rigatti, L.; Wang, H.; Franicola, D.; Sivanathan, A.; Greenberger, J.S. Evolution of malignant plasmacytoma cell lines from K14E7 Fancd2-/- mouse long-term bone marrow cultures. Oncotarget 2016, 7, 68449–68472. [Google Scholar] [CrossRef] [Green Version]
- Kasai, F.; Hirayama, N.; Ozawa, M.; Iemura, M.; Kohara, A. Changes of heterogeneous cell populations in the Ishikawa cell line during long-term culture: Proposal for an in vitro clonal evolution model of tumor cells. Genomics 2016, 107, 259–266. [Google Scholar] [CrossRef]
- Missiaglia, E.; Selfe, J.; Hamdi, M.; Williamson, D.; Schaaf, G.; Fang, C.; Koster, J.; Summersgill, B.; Messahel, B.; Versteeg, R.; et al. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: An approach to identify candidate genes involved in tumor development. Genes Chromosom. Cancer 2009, 48, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Muff, R.; Rath, P.; Kumar, R.M.R.; Husmann, K.; Born, W.; Baudis, M.; Fuchs, B. Genomic instability of osteosarcoma cell lines in culture: Impact on the prediction of metastasis relevant genes. PLoS ONE 2015, 10, e0125611. [Google Scholar] [CrossRef] [Green Version]
- Bairoch, A. The cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.H.; Huang, J.; Bi, Y.; Su, Y.; Tang, Y.; He, B.C.; He, Y.; Luo, J.; Wang, Y.; Chen, L.; et al. Activation of RXR and RAR signaling promotes myogenic differentiation of myoblastic C2C12 cells. Differentiation 2009, 78, 195–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barlow, J.W.; Wiley, J.C.; Mous, M.; Narendran, A.; Gee, M.F.W.; Goldberg, M.; Sexsmith, E.; Malkin, D. Differentiation of rhabdomyosarcoma cell lines using retinoic acid. Pediatr. Blood Cancer 2006, 47, 773–784. [Google Scholar] [CrossRef]
- Al-Tahan, A.; Sarkis, O.; Harajly, M.; Baghdadi, O.K.; Zibara, K.; Boulos, F.; Dighe, D.; Kregel, S.; Bazarbachi, A.; El-Sabban, M.; et al. Retinoic acid fails to induce cell cycle arrest with myogenic differentiation in rhabdomyosarcoma. Pediatr. Blood Cancer 2012, 58, 877–884. [Google Scholar] [CrossRef]
- Gryder, B.E.; Yohe, M.E.; Chou, H.C.; Zhang, X.; Marques, J.; Wachtel, M.; Schaefer, B.; Sen, N.; Song, Y.; Gualtieri, A.; et al. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 2017, 7, 884–899. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Runa, F.; Hamalian, S.; Meade, K.; Shisgal, P.; Gray, P.C.; Kelber, J.A. Tumor microenvironment heterogeneity: Challenges and opportunities. Curr. Mol. Biol. Rep. 2017, 3, 218–229. [Google Scholar] [CrossRef]
- Zyryanova, T.; Schneider, R.; Adams, V.; Sittig, D.; Kerner, C.; Gebhardt, C.; Ruffert, H.; Glasmacher, S.; Hepp, P.; Punkt, K.; et al. Skeletal muscle expression of the adhesion-GPCR CD97: CD97 deletion induces an abnormal structure of the sarcoplasmatic reticulum but does not impair skeletal muscle function. PLoS ONE 2014, 9, e100513. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.I.; Abraham, J.; Mansoor, A.; Druker, B.J.; Tyner, J.W.; Keller, C. PDGFRβ reverses EphB4 signaling in alveolar rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 2014, 111, 6383–6388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shipley, J.; Martins, A.S.; Olmos, D.; Missiaglia, E. Targeting the insulin-like growth factor pathway in rhabdomyosarcomas: Rationale and future perspectives. Sarcoma 2011, 2011, 11. [Google Scholar]
- Darvishi, B.; Boroumandieh, S.; Majidzadeh-A, K.; Salehi, M.; Jafari, F.; Farahmand, L. The role of activated leukocyte cell adhesion molecule (ALCAM) in cancer progression, invasion, metastasis and recurrence: A novel cancer stem cell marker and tumor-specific prognostic marker. Exp. Mol. Pathol. 2020, 115, 104443. [Google Scholar] [CrossRef] [PubMed]
- Read, T.A.; Fogarty, M.P.; Markant, S.L.; McLendon, R.E.; Wei, Z.; Ellison, D.W.; Febbo, P.G.; Wechsler-Reya, R.J. Identification of CD15 as a Marker for Tumor-Propagating Cells in a Mouse Model of Medulloblastoma. Cancer Cell 2009, 15, 135–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziprin, P.; Alkhamesi, N.A.; Ridgway, P.F.; Peck, D.H.; Darzi, A.W. Tumour-expressed CD43 (sialophorin) mediates tumour-mesothelial cell adhesion. Biol. Chem. 2004, 385, 755–761. [Google Scholar] [CrossRef]
- Kadaja-Saarepuu, L.; Laos, S.; Jääger, K.; Viil, J.; Balikova, A.; Lõoke, M.; Hansson, G.C.; Maimets, T. CD43 promotes cell growth and helps to evade FAS-mediated apoptosis in non-hematopoietic cancer cells lacking the tumor suppressors p53 or ARF. Oncogene 2008, 27, 1705–1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wang, Y.; Rong, S.K.; Li, L.; Chen, T.; Fan, Y.Y.; Wang, Y.F.; Yang, C.R.; Yang, C.; Cho, W.C.; et al. Integrin α1 promotes tumorigenicity and progressive capacity of colorectal cancer. Int. J. Biol. Sci. 2020, 16, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Gharibi, A.; La Kim, S.; Molnar, J.; Brambilla, D.; Adamian, Y.; Hoover, M.; Hong, J.; Lin, J.; Wolfenden, L.; Kelber, J.A. ITGA1 is a pre-malignant biomarker that promotes therapy resistance and metastatic potential in pancreatic cancer. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Okazaki, K.; Nakayama, Y.; Shibao, K.; Hirata, K.; Nagata, N.; Itoh, H. Enhancement of metastatic activity of colon cancer as influenced by expression of cell surface antigens. J. Surg. Res. 1998, 78, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Adorno-Cruz, V.; Liu, H. Regulation and functions of integrin α2 in cell adhesion and disease. Genes Dis. 2019, 6, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Varzavand, A.; Hacker, W.; Ma, D.; Gibson-Corley, K.; Hawayek, M.; Tayh, O.J.; Brown, J.A.; Henry, M.D.; Stipp, C.S. α3β1 integrin suppresses prostate cancer metastasis via regulation of the Hippo pathway. Cancer Res. 2016, 76, 6577–6587. [Google Scholar] [CrossRef] [Green Version]
- Du, Q.; Wang, W.; Liu, T.; Shang, C.; Huang, J.; Liao, Y.; Qin, S.; Chen, Y.; Liu, P.; Liu, J.; et al. High Expression of Integrin α3 Predicts Poor Prognosis and Promotes Tumor Metastasis and Angiogenesis by Activating the c-Src/Extracellular Signal-Regulated Protein Kinase/Focal Adhesion Kinase Signaling Pathway in Cervical Cancer. Front. Oncol. 2020, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Benedicto, A.; Romayor, I.; Arteta, B. Role of liver ICAM-1 in metastasis. Oncol. Lett. 2017, 14, 3883–3892. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Xu, L.; Feng, M.; Wang, W. Role of CD97 small isoform in human cervical carcinoma. Int. J. Exp. Pathol. 2019, 100, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Trojanowicz, B.; Ye, L.; Li, C.; Zhang, L.; Li, X.; Li, G.; Zheng, Y.; Chen, L. The invasion and metastasis promotion role of CD97 small isoform in gastric carcinoma. PLoS ONE 2012, 7, e39989. [Google Scholar] [CrossRef]
- Tanase, C.; Gheorghisan-Galateanu, A.A.; Popescu, I.D.; Mihai, S.; Codrici, E.; Albulescu, R.; Hinescu, M.E. Cd36 and cd97 in pancreatic cancer versus other malignancies. Int. J. Mol. Sci. 2020, 21, 5656. [Google Scholar] [CrossRef]
- Yin, Y.; Xu, X.; Tang, J.; Zhang, W.; Zhangyuan, G.; Ji, J.; Deng, L.; Lu, S.; Zhuo, H.; Sun, B. CD97 Promotes Tumor Aggressiveness Through the Traditional G Protein–Coupled Receptor–Mediated Signaling in Hepatocellular Carcinoma. Hepatology 2018, 68, 1865–1878. [Google Scholar] [CrossRef] [Green Version]
- Yoon, K.J.; Miller, A.L.; Kreitzburg, K.M. The role of ICAM-2 in neuroblastoma. Oncoscience 2015, 2, 915–916. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Tamura, M.; Takeda, K.; Ogi, K.; Nakagaki, T.; Koyama, R.; Idogawa, M.; Hiratsuka, H.; Tokino, T. Identification and characterization of the intercellular adhesion molecule-2 gene as a novel p53 target. Oncotarget 2016, 7, 61426–61437. [Google Scholar] [CrossRef] [Green Version]
- Steller, E.J.A.; Raats, D.A.; Koster, J.; Rutten, B.; Govaert, K.M.; Emmink, B.L.; Snoeren, N.; van Hooff, S.R.; Holstege, F.C.P.; Maas, C.; et al. PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia 2013, 15, 204–217. [Google Scholar] [CrossRef] [Green Version]
- Heldin, C.H. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun. Signal. 2013, 11, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.K.; Ham, M.H.; Lee, S.Y.; Shin, M.J.; Kim, Y.E.; Song, P.; Suh, D.S.; Kim, J.H. CD166 promotes the cancer stem-like properties of primary epithelial ovarian cancer cells. BMB Rep. 2020, 53, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Devis, L.; Moiola, C.P.; Masia, N.; Martinez-Garcia, E.; Santacana, M.; Stirbat, T.V.; Brochard-Wyart, F.; García, Á.; Alameda, F.; Cabrera, S.; et al. Activated leukocyte cell adhesion molecule (ALCAM) is a marker of recurrence and promotes cell migration, invasion, and metastasis in early-stage endometrioid endometrial cancer. J. Pathol. 2017, 241, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wang, J.; Ma, L.; Tang, X.; Qiao, Y.; Pan, Q.; Yu, Y.; Sun, F. CD166 plays a pro-carcinogenic role in liver cancer cells via inhibition of FOXO proteins through AKT. Oncol. Rep. 2014, 32, 677–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jezierska, A.; Matysiak, W.; Motyl, T. ALCAM/CD166 protects breast cancer cells against apoptosis and autophagy. Med. Sci. Monit. 2006, 12, BR263–BR273. [Google Scholar] [PubMed]
- Swart, G.W.M.; Lunter, P.C.; Van Kilsdonk, J.W.J.; Van Kempen, L.C.L.T. Activated leukocyte cell adhesion molecule (ALCAM/CD166): Signaling at the divide of melanoma cell clustering and cell migration? Cancer Metastasis Rev. 2005, 24, 223–236. [Google Scholar] [CrossRef]
- Zhang, H.; Fagan, D.H.; Zeng, X.; Freeman, K.T.; Sachdev, D.; Yee, D. Inhibition of cancer cell proliferation and metastasis by insulin receptor downregulation. Oncogene 2010, 29, 2517–2527. [Google Scholar] [CrossRef] [Green Version]
- Vigneri, R.; Goldfine, I.D.; Frittitta, L. Insulin, insulin receptors, and cancer. J. Endocrinol. Invest. 2016, 39, 1365–1376. [Google Scholar] [CrossRef]
- Belfiore, A. The Role of Insulin Receptor Isoforms and Hybrid Insulin/IGF-I Receptors in Human Cancer. Curr. Pharm. Des. 2007, 13, 671–686. [Google Scholar] [CrossRef] [PubMed]
- Belyea, B.C.; Naini, S.; Bentley, R.C.; Linardic, C.M. Inhibition of the notch-hey1 axis blocks embryonal rhabdomyosarcoma tumorigenesis. Clin. Cancer Res. 2011, 17, 7324–7336. [Google Scholar] [CrossRef] [Green Version]
- Hinson, A.R.P.; Jones, R.; Lisa, L.E.; Belyea, B.C.; Barr, F.G.; Linardic, C.M. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: Utility and pitfalls. Front. Oncol. 2013, 3, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masters, J.R. Cell-line authentication: End the scandal of false cell lines. Nature 2012, 492, 186. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Nishimura, R.; Yoshida, K.; Shimamura, T.; Shiraishi, Y.; Sato, Y.; Kato, M.; Chiba, K.; Tanaka, H.; Hoshino, N.; et al. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat. Commun. 2015, 6, 7557. [Google Scholar] [CrossRef] [Green Version]
- Chiappalupi, S.; Riuzzi, F.; Fulle, S.; Donato, R.; Sorci, G. Defective RAGE activity in embryonal rhabdomyosarcoma cells results in high PAX7 levels that sustain migration and invasiveness. Carcinogenesis 2014, 35, 2382–2392. [Google Scholar] [CrossRef] [Green Version]
- Zammit, P.S.; Relaix, F.; Nagata, Y.; Ruiz, A.P.; Collins, C.A.; Partridge, T.A.; Beauchamp, J.R. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006, 119, 1824–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riuzzi, F.; Sorci, G.; Sagheddu, R.; Sidoni, A.; Alaggio, R.; Ninfo, V.; Donato, R. RAGE signaling deficiency in rhabdomyosarcoma cells causes upregulation of PAX7 and uncontrolled proliferation. J. Cell Sci. 2014, 127, 1699–1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Benezra, R.; Sassoon, D.A. Id expression during mouse development: A role in morphogenesis. Dev. Dyn. 1992, 194, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Tonin, P.N.; Scrable, H.; Shimada, H.; Cavenee, W.K. Muscle-specific Gene Expression in Rhabdomyosarcomas and Stages of Human Fetal Skeletal Muscle Development. Cancer Res. 1991, 51, 5100–5106. [Google Scholar] [PubMed]
- Lasorella, A.; Benezra, R.; Iavarone, A. The ID proteins: Master regulators of cancer stem cells and tumour aggressiveness. Nat. Rev. Cancer 2014, 14, 77–91. [Google Scholar] [CrossRef]
- Buford, T.W.; Cooke, M.B.; Shelmadine, B.D.; Hudson, G.M.; Redd, L.L.; Willoughby, D.S. Differential gene expression of FoxO1, ID1, and ID3 between young and older men and associations with muscle mass and function. Aging Clin. Exp. Res. 2011, 23, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Dawson, L.E.; D’Agostino, L.; Hakim, A.A.; Lackman, R.D.; Brown, S.A.; Sensenig, R.B.; Antonello, Z.A.; Kuzin, I.I. Induction of Myogenic Differentiation Improves Chemosensitivity of Chemoresistant Cells in Soft-Tissue Sarcoma Cell Lines. Sarcoma 2020, 2020, 8647981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zibat, A.; Missiaglia, E.; Rosenberger, A.; Pritchard-Jones, K.; Shipley, J.; Hahn, H.; Fulda, S. Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene 2010, 29, 6323–6330. [Google Scholar] [CrossRef] [Green Version]
- Tenente, I.M.; Hayes, M.N.; Ignatius, M.S.; McCarthy, K.; Yohe, M.; Sindiri, S.; Gryder, B.; Oliveira, M.L.; Ramakrishnan, A.; Tang, Q.; et al. Myogenic regulatory transcription factors regulate growth in rhabdomyosarcoma. eLife 2017, 6, e19214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charytonowicz, E.; Cordon-Cardo, C.; Matushansky, I.; Ziman, M. Alveolar rhabdomyosarcoma: Is the cell of origin a mesenchymal stem cell? Cancer Lett. 2009, 279, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.P.; Nishijo, K.; Chen, H.-I.H.; Yi, X.; Schuetze, D.P.; Pal, R.; Prajapati, S.I.; Abraham, J.; Arenkiel, B.R.; Chen, Q.-R.; et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell 2011, 19, 177–191. [Google Scholar] [CrossRef] [Green Version]
- Charville, G.W.; Varma, S.; Forgó, E.; Dumont, S.N.; Zambrano, E.; Trent, J.C.; Lazar, A.J.; Van De Rijn, M. PAX7 expression in rhabdomyosarcoma, related soft tissue tumors, and small round blue cell neoplasms. Am. J. Surg. Pathol. 2016, 40, 1305–1315. [Google Scholar] [CrossRef] [Green Version]
- Tiffin, N.; Williams, R.D.; Shipley, J.; Pritchard-Jones, K. PAX7 expression in embryonal rhabdomyosarcoma suggests an origin in muscle satellite cells. Br. J. Cancer 2003, 89, 327–332. [Google Scholar] [CrossRef] [Green Version]
- Tomescu, O.; Xia, S.J.; Strezlecki, D.; Bennicelli, J.L.; Ginsberg, J.; Pawel, B.; Barr, F.G. Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Lab. Investig. 2004, 84, 1060–1070. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, C. Identification of a new class of PAX3-FKHR target promoters: A role of the Pax3 paired box DNA binding domain. Oncogene 2007, 26, 1595–1605. [Google Scholar] [CrossRef] [Green Version]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Hangan, D.; Uniyal, S.; Morris, V.L.; MacDonald, I.C.; von Ballestrem, C.; Chau, T.; Schmidt, E.E.; Chambers, A.F.; Groom, A.C.; Chan, B.M. Integrin VLA-2 (alpha2beta1) function in postextravasation movement of human rhabdomyosarcoma RD cells in the liver. Cancer Res. 1996, 56, 3142–3149. [Google Scholar]









| Loci | RH30 from ATCC | RH30 Donated | RH30 Data from the Expasy Database |
|---|---|---|---|
| D3S1358 | 15 | 15 | 15 |
| vWA | 17; 18 | 17; 18 | 17; 18 |
| D16S539 | 12 | 12 | 12 |
| D2S1338 | 17; 20 | 17; 20 | 17; 20 |
| Amelogenin | X; Y | X | X (COG) or X; Y (ATCC) |
| D8S1179 | 12; 15 | 12; 15 | 12; 15 |
| D21S11 | 29; 31.2 | 29; 31.2 | 29, 31.2 |
| D18S51 | 15; 16 | 15; 16 | 15; 16 |
| D19S433 | 14; 15.2 | 14; 15.2 | 14; 15.2 |
| THO1 | 9; 9.3 | 9; 9.3 | 9; 9.3 |
| FGA | 22 | 22 | 22 |
| Marker | Other Names | RH30 PAX7− [%] | RH30 PAX7+ [%] | Significance in Tumor Biology |
|---|---|---|---|---|
| CD15 | SSEA-1 | 68.97 | 3.61 | Involved in tumor propagation; its expression increases proliferation in vitro and in vivo [40]. |
| CD43 | Sialophorin | 67.99 | 3.05 | Promotes cell growth by increasing cell survival, viability and colony formation; helps to evade FAS-mediated apoptosis [42]; ligand for ICAM-1—role in tumor cell-peritoneal adhesion [41]. |
| CD49a | VLA-1; Integrin α1 | 36.25 | 96.24 | Cell-surface receptor for collagens and laminins; involved in cell–cell adhesion and plays a role in inflammation and fibrosis; significantly upregulated in colorectal tumors thus promotes tumorigenicity and progressive capacity of this type of cancer [43]; associated with an invasive/metastatic phenotype; frequently overexpressed in malignancies such as melanoma, prostate, bladder, liver myeloma; a pre-malignant biomarker in pancreatic cancer [44]. |
| CD49b | VLA-2; Integrin α2 | 4.33 | 85.29 | It binds only to the β1 subunit (CD29). Involved in the metastatic activity and cell adhesion to an extracellular matrix (ECM) [45]; it participates in cell motility, angiogenesis and cell stemness; the role of integrin α2 in cancers is still not well understood and varies according to cancer type, i.e., angiogenesis effect [46]. |
| CD49c | VL3A; Integrin α3 | 31.69 | 98.71 | Involved in the increase of metastatic activity and promotion of adhesion to an extracellular matrix (ECM) [45]; may acts as pro-tumoral or tumoricidal factor depends on types of cancer, e.g., suppresses the metastasis of prostate cancer [47], whereas promotes migration and invasion of cervical cancer cells [48]. |
| CD54 | ICAM-1 | 4.25 | 96.04 | Determines malignant potential of cancer via promotion of extraluminal migration of tumor cells [41]; key role during tumor progression and metastasis formation; involved in the activation of pro-inflammatory cascades, and the mediation of multiple signaling pathways that regulate metastasis, such as tumor cell adhesion and transmigration, immune escape, desmoplasia, and angiogenesis [49]. |
| CD97 | Adhesion G protein-coupled receptor E5 | 45.89 | 73.09 | Elevated expression is associated with the dedifferentiation, aggressiveness, metastasis and invasion of tumor [50,51]; induced or upregulated, and/or biochemically modified in various malignancies. Involved in cell adhesion, migration, invasiveness, and regulation of intercellular junctions; affects tumor aggressiveness through stimulation of its ligand CD55 [52,53]. |
| CD102 | ICAM-2 | 0.01 | 23.72 | Role in cellular adhesion; via interactions with extracellular matrix proteins plays a role in cell motility, i.e., might inhibit tumor cell motility and suppresses the metastatic potential [54]; ICAM-2 is underexpressed in human cancer tissues with p53 mutation; its low expression is associated with poor survival in patients with various cancers. Induction of ICAM-2 by p53 has a key role in inhibiting migration and invasion [55]. |
| CD140b | PDGFRB | 3.89 | 52.13 | Promotes metastasis; tumor cells may acquire PDGFRB expression following epithelial-mesenchymal transition (EMT) during metastasis; it may contribute to form the aggressive phenotype of colorectal tumors with mesenchymal properties [56]; overexpression may lead to tumor cell growth and promote tumorigenesis; associated with certain malignant and non-malignant diseases characterised by extensive proliferation [57]. |
| CD166 | ALCAM | 0.64 | 71.96 | CD166 positive cells exhibit some CSCs-like (cancer stem cells) properties, such as sphere-forming ability, cell migration, adhesion, and high tumorigenic potential in vivo [58]; marker has been identified in many types of cancers; indicated as a biomarker for ovarian CSCs [58]; promotes cell migration, invasion, and metastasis in early-stage endometrial cancer [59]; plays a procarcinogenic role in liver cancer cells [60]; CD166 expression is positively correlated with the progression of breast cancer and melanoma [61,62]. |
| CD220 | Insulin receptor (IR) | 20.08 | 59.36 | Downregulation of the receptor inhibits cancer cell proliferation, angiogenesis and metastasis; IRs are highly expressed in malignant cells [63]; in many cancer cells, A isoform is more predominant form than isoform B; IRs have mitogenic effect promoting cancer growth [64]. |
| CD221 | IGF-1R | 42.25 | 98.37 | IGF-1R forms hybrid receptors with the isoform A of IR (IR-A), which are commonly overexpressed in human malignancies; hybrid receptors may be regard as potential targets of anti-cancer therapy; the inhibition of IGF-1R/IR-A activity may block cancer growth and metastatic spread [65]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skrzypek, K.; Adamek, G.; Kot, M.; Badyra, B.; Majka, M. Progression and Differentiation of Alveolar Rhabdomyosarcoma Is Regulated by PAX7 Transcription Factor—Significance of Tumor Subclones. Cells 2021, 10, 1870. https://doi.org/10.3390/cells10081870
Skrzypek K, Adamek G, Kot M, Badyra B, Majka M. Progression and Differentiation of Alveolar Rhabdomyosarcoma Is Regulated by PAX7 Transcription Factor—Significance of Tumor Subclones. Cells. 2021; 10(8):1870. https://doi.org/10.3390/cells10081870
Chicago/Turabian StyleSkrzypek, Klaudia, Grażyna Adamek, Marta Kot, Bogna Badyra, and Marcin Majka. 2021. "Progression and Differentiation of Alveolar Rhabdomyosarcoma Is Regulated by PAX7 Transcription Factor—Significance of Tumor Subclones" Cells 10, no. 8: 1870. https://doi.org/10.3390/cells10081870