Plasmacytoid Dendritic Cells, a Novel Target in Myeloid Neoplasms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Biology and Functions of pDCs
2.1. Definition of pDC Subset
2.2. Development of pDCs
2.3. Transcriptomic Network of pDC Development
2.4. Innate Activation of pDC
2.5. Non-Canonical pDCs
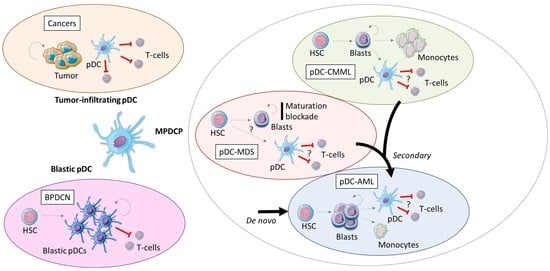
3. Blastic pDC Neoplasm
4. Tumor-Infiltrating pDCs in Solid Cancers
5. Mature pDCs Proliferation in Myeloid Neoplasms
5.1. Mature pDCs Proliferation in CMML
5.2. Mature pDCs Proliferation in MDS
5.3. Mature pDC Proliferation in AML
6. Current Therapies and Perspective in pDC-AML
6.1. Current Therapies
6.2. Targeted pDC from pDC-AML
6.3. Targeted RUNX1 Mutations and pDC
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Swiecki, M.; Colonna, M. The Multifaceted Biology of Plasmacytoid Dendritic Cells. Nat. Rev. Immunol. 2015, 15, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; IARC Publications: Lyon, France, 2017; ISBN 978-92-832-4494-3. [Google Scholar]
- Xiao, W.; Chan, A.; Waarts, M.R.; Mishra, T.; Liu, Y.; Cai, S.F.; Yao, J.; Gao, Q.; Bowman, R.L.; Koche, R.P.; et al. Plasmacytoid Dendritic Cell Expansion Defines a Distinct Subset of RUNX1-Mutated Acute Myeloid Leukemia. Blood 2021, 137, 1377–1391. [Google Scholar] [CrossRef]
- Zalmaï, L.; Viailly, P.-J.; Biichle, S.; Cheok, M.; Soret, L.; Angelot-Delettre, F.; Petrella, T.; Collonge-Rame, M.-A.; Seilles, E.; Geffroy, S.; et al. Plasmacytoid Dendritic Cells Proliferation Associated with Acute Myeloid Leukemia: Phenotype Profile and Mutation Landscape. Haematologica 2021, 106, 3056–3066. [Google Scholar] [CrossRef] [PubMed]
- Giovanelli, P.; Sandoval, T.A.; Cubillos-Ruiz, J.R. Dendritic Cell Metabolism and Function in Tumors. Trends Immunol. 2019, 40, 699–718. [Google Scholar] [CrossRef] [PubMed]
- Reizis, B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 2019, 50, 37–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cella, M.; Jarrossay, D.; Facchetti, F.; Alebardi, O.; Nakajima, H.; Lanzavecchia, A.; Colonna, M. Plasmacytoid Monocytes Migrate to Inflamed Lymph Nodes and Produce Large Amounts of Type I Interferon. Nat. Med. 1999, 5, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Siegal, F.P.; Kadowaki, N.; Shodell, M.; Fitzgerald-Bocarsly, P.A.; Shah, K.; Ho, S.; Antonenko, S.; Liu, Y.J. The Nature of the Principal Type 1 Interferon-Producing Cells in Human Blood. Science 1999, 284, 1835–1837. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, P.F.; Tussiwand, R. Novel Concepts in Plasmacytoid Dendritic Cell (PDC) Development and Differentiation. Mol. Immunol. 2020, 126, 25–30. [Google Scholar] [CrossRef]
- Summers, K.L.; Hock, B.D.; McKenzie, J.L.; Hart, D.N. Phenotypic Characterization of Five Dendritic Cell Subsets in Human Tonsils. Am. J. Pathol. 2001, 159, 285–295. [Google Scholar] [CrossRef] [Green Version]
- Colonna, M.; Trinchieri, G.; Liu, Y.-J. Plasmacytoid Dendritic Cells in Immunity. Nat. Immunol. 2004, 5, 1219–1226. [Google Scholar] [CrossRef]
- Liu, Y.-J. IPC: Professional Type 1 Interferon-Producing Cells and Plasmacytoid Dendritic Cell Precursors. Annu. Rev. Immunol. 2005, 23, 275–306. [Google Scholar] [CrossRef]
- Sathe, P.; Vremec, D.; Wu, L.; Corcoran, L.; Shortman, K. Convergent Differentiation: Myeloid and Lymphoid Pathways to Murine Plasmacytoid Dendritic Cells. Blood 2013, 121, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Naik, S.H.; Sathe, P.; Park, H.-Y.; Metcalf, D.; Proietto, A.I.; Dakic, A.; Carotta, S.; O’Keeffe, M.; Bahlo, M.; Papenfuss, A.; et al. Development of Plasmacytoid and Conventional Dendritic Cell Subtypes from Single Precursor Cells Derived In Vitro and In Vivo. Nat. Immunol. 2007, 8, 1217–1226. [Google Scholar] [CrossRef]
- Onai, N.; Kurabayashi, K.; Hosoi-Amaike, M.; Toyama-Sorimachi, N.; Matsushima, K.; Inaba, K.; Ohteki, T. A Clonogenic Progenitor with Prominent Plasmacytoid Dendritic Cell Developmental Potential. Immunity 2013, 38, 943–957. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Chow, K.V.; Soo, P.; Xu, Z.; Brady, J.L.; Lawlor, K.E.; Masters, S.L.; O’keeffe, M.; Shortman, K.; Zhang, J.-G.; et al. Plasmacytoid Dendritic Cells Are Short-Lived: Reappraising the Influence of Migration, Genetic Factors and Activation on Estimation of Lifespan. Sci. Rep. 2016, 6, 25060. [Google Scholar] [CrossRef] [Green Version]
- Kohara, H.; Omatsu, Y.; Sugiyama, T.; Noda, M.; Fujii, N.; Nagasawa, T. Development of Plasmacytoid Dendritic Cells in Bone Marrow Stromal Cell Niches Requires CXCL12-CXCR4 Chemokine Signaling. Blood 2007, 110, 4153–4160. [Google Scholar] [CrossRef]
- Zou, W.; Machelon, V.; Coulomb-L’Hermin, A.; Borvak, J.; Nome, F.; Isaeva, T.; Wei, S.; Krzysiek, R.; Durand-Gasselin, I.; Gordon, A.; et al. Stromal-Derived Factor-1 in Human Tumors Recruits and Alters the Function of Plasmacytoid Precursor Dendritic Cells. Nat. Med. 2001, 7, 1339–1346. [Google Scholar] [CrossRef]
- Nutt, S.L.; Chopin, M. Transcriptional Networks Driving Dendritic Cell Differentiation and Function. Immunity 2020, 52, 942–956. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Chen, T.-T.; Pai, L.-M.; Wesoly, J.; Bluyssen, H.A.R.; Lee, C.-K. A Type I IFN-Flt3 Ligand Axis Augments Plasmacytoid Dendritic Cell Development from Common Lymphoid Progenitors. J. Exp. Med. 2013, 210, 2515–2522. [Google Scholar] [CrossRef]
- Sathaliyawala, T.; O’Gorman, W.E.; Greter, M.; Bogunovic, M.; Konjufca, V.; Hou, Z.E.; Nolan, G.P.; Miller, M.J.; Merad, M.; Reizis, B. Mammalian Target of Rapamycin Controls Dendritic Cell Development Downstream of Flt3 Ligand Signaling. Immunity 2010, 33, 597–606. [Google Scholar] [CrossRef] [Green Version]
- Esashi, E.; Wang, Y.-H.; Perng, O.; Qin, X.-F.; Liu, Y.-J.; Watowich, S.S. The Signal Transducer STAT5 Inhibits Plasmacytoid Dendritic Cell Development by Suppressing Transcription Factor IRF8. Immunity 2008, 28, 509–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carotta, S.; Dakic, A.; D’Amico, A.; Pang, S.H.M.; Greig, K.T.; Nutt, S.L.; Wu, L. The Transcription Factor PU.1 Controls Dendritic Cell Development and Flt3 Cytokine Receptor Expression in a Dose-Dependent Manner. Immunity 2010, 32, 628–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cisse, B.; Caton, M.L.; Lehner, M.; Maeda, T.; Scheu, S.; Locksley, R.; Holmberg, D.; Zweier, C.; den Hollander, N.S.; Kant, S.G.; et al. Transcription Factor E2-2 Is an Essential and Specific Regulator of Plasmacytoid Dendritic Cell Development. Cell 2008, 135, 37–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagasawa, M.; Schmidlin, H.; Hazekamp, M.G.; Schotte, R.; Blom, B. Development of Human Plasmacytoid Dendritic Cells Depends on the Combined Action of the Basic Helix-Loop-Helix Factor E2-2 and the Ets Factor Spi-B. Eur. J. Immunol. 2008, 38, 2389–2400. [Google Scholar] [CrossRef]
- Upadhaya, S.; Sawai, C.M.; Papalexi, E.; Rashidfarrokhi, A.; Jang, G.; Chattopadhyay, P.; Satija, R.; Reizis, B. Kinetics of Adult Hematopoietic Stem Cell Differentiation in vivo. J. Exp. Med. 2018, 215, 2815–2832. [Google Scholar] [CrossRef] [Green Version]
- Grajkowska, L.T.; Ceribelli, M.; Lau, C.M.; Warren, M.E.; Tiniakou, I.; Nakandakari Higa, S.; Bunin, A.; Haecker, H.; Mirny, L.A.; Staudt, L.M.; et al. Isoform-Specific Expression and Feedback Regulation of E Protein TCF4 Control Dendritic Cell Lineage Specification. Immunity 2017, 46, 65–77. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, H.S.; Ceribelli, M.; Matos, I.; Lazarovici, A.; Bussemaker, H.J.; Lasorella, A.; Hiebert, S.W.; Liu, K.; Staudt, L.M.; Reizis, B. ETO Family Protein Mtg16 Regulates the Balance of Dendritic Cell Subsets by Repressing Id2. J. Exp. Med. 2014, 211, 1623–1635. [Google Scholar] [CrossRef] [Green Version]
- Ippolito, G.C.; Dekker, J.D.; Wang, Y.-H.; Lee, B.-K.; Shaffer, A.L.; Lin, J.; Wall, J.K.; Lee, B.-S.; Staudt, L.M.; Liu, Y.-J.; et al. Dendritic Cell Fate Is Determined by BCL11A. Proc. Natl. Acad. Sci. USA 2014, 111, E998-1006. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Satpathy, A.T.; Kc, W.; Liu, P.; Murphy, T.L.; Murphy, K.M. Bcl11a Controls Flt3 Expression in Early Hematopoietic Progenitors and Is Required for PDC Development in Vivo. PLoS ONE 2013, 8, e64800. [Google Scholar] [CrossRef] [Green Version]
- Scott, C.L.; Soen, B.; Martens, L.; Skrypek, N.; Saelens, W.; Taminau, J.; Blancke, G.; Van Isterdael, G.; Huylebroeck, D.; Haigh, J.; et al. The Transcription Factor Zeb2 Regulates Development of Conventional and Plasmacytoid DCs by Repressing Id2. J. Exp. Med. 2016, 213, 897–911. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Briseño, C.G.; Grajales-Reyes, G.E.; Haldar, M.; Iwata, A.; Kretzer, N.M.; Kc, W.; Tussiwand, R.; Higashi, Y.; Murphy, T.L.; et al. Transcription Factor Zeb2 Regulates Commitment to Plasmacytoid Dendritic Cell and Monocyte Fate. Proc. Natl. Acad. Sci. USA 2016, 113, 14775–14780. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, I.; Hoshino, K.; Sugiyama, T.; Yamazaki, C.; Yano, T.; Iizuka, A.; Hemmi, H.; Tanaka, T.; Saito, M.; Sugiyama, M.; et al. Spi-B Is Critical for Plasmacytoid Dendritic Cell Function and Development. Blood 2012, 120, 4733–4743. [Google Scholar] [CrossRef] [Green Version]
- Sawai, C.M.; Sisirak, V.; Ghosh, H.S.; Hou, E.Z.; Ceribelli, M.; Staudt, L.M.; Reizis, B. Transcription Factor Runx2 Controls the Development and Migration of Plasmacytoid Dendritic Cells. J. Exp. Med. 2013, 210, 2151–2159. [Google Scholar] [CrossRef] [Green Version]
- Chopin, M.; Preston, S.P.; Lun, A.T.L.; Tellier, J.; Smyth, G.K.; Pellegrini, M.; Belz, G.T.; Corcoran, L.M.; Visvader, J.E.; Wu, L.; et al. RUNX2 Mediates Plasmacytoid Dendritic Cell Egress from the Bone Marrow and Controls Viral Immunity. Cell Rep. 2016, 15, 866–878. [Google Scholar] [CrossRef] [Green Version]
- Honda, K.; Yanai, H.; Negishi, H.; Asagiri, M.; Sato, M.; Mizutani, T.; Shimada, N.; Ohba, Y.; Takaoka, A.; Yoshida, N.; et al. IRF-7 Is the Master Regulator of Type-I Interferon-Dependent Immune Responses. Nature 2005, 434, 772–777. [Google Scholar] [CrossRef]
- Yasuda, K.; Nündel, K.; Watkins, A.A.; Dhawan, T.; Bonegio, R.G.; Ubellacker, J.M.; Marshak-Rothstein, A.; Rifkin, I.R. Phenotype and Function of B Cells and Dendritic Cells from Interferon Regulatory Factor 5-Deficient Mice with and without a Mutation in DOCK2. Int. Immunol. 2013, 25, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Sichien, D.; Scott, C.L.; Martens, L.; Vanderkerken, M.; Van Gassen, S.; Plantinga, M.; Joeris, T.; De Prijck, S.; Vanhoutte, L.; Vanheerswynghels, M.; et al. IRF8 Transcription Factor Controls Survival and Function of Terminally Differentiated Conventional and Plasmacytoid Dendritic Cells, Respectively. Immunity 2016, 45, 626–640. [Google Scholar] [CrossRef] [Green Version]
- Bao, M.; Wang, Y.; Liu, Y.; Shi, P.; Lu, H.; Sha, W.; Weng, L.; Hanabuchi, S.; Qin, J.; Plumas, J.; et al. NFATC3 Promotes IRF7 Transcriptional Activity in Plasmacytoid Dendritic Cells. J. Exp. Med. 2016, 213, 2383–2398. [Google Scholar] [CrossRef]
- Kim, T.W.; Hong, S.; Lin, Y.; Murat, E.; Joo, H.; Kim, T.; Pascual, V.; Liu, Y.-J. Transcriptional Repression of IFN Regulatory Factor 7 by MYC Is Critical for Type I IFN Production in Human Plasmacytoid Dendritic Cells. J. Immunol. 2016, 197, 3348–3359. [Google Scholar] [CrossRef] [Green Version]
- Mastio, J.; Simand, C.; Cova, G.; Kastner, P.; Chan, S.; Kirstetter, P. Ikaros Cooperates with Notch Activation and Antagonizes TGFβ Signaling to Promote PDC Development. PLoS Genet. 2018, 14, e1007485. [Google Scholar] [CrossRef]
- Ma, S.; Wan, X.; Deng, Z.; Shi, L.; Hao, C.; Zhou, Z.; Zhou, C.; Fang, Y.; Liu, J.; Yang, J.; et al. Epigenetic Regulator CXXC5 Recruits DNA Demethylase Tet2 to Regulate TLR7/9-Elicited IFN Response in PDCs. J. Exp. Med. 2017, 214, 1471–1491. [Google Scholar] [CrossRef]
- Gong, T.; Liu, L.; Jiang, W.; Zhou, R. DAMP-Sensing Receptors in Sterile Inflammation and Inflammatory Diseases. Nat. Rev. Immunol. 2020, 20, 95–112. [Google Scholar] [CrossRef]
- Janeway, C.A. Approaching the Asymptote? Evolution and Revolution in Immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54, 1–13. [Google Scholar] [CrossRef]
- Matzinger, P. Tolerance, Danger, and the Extended Family. Annu. Rev. Immunol. 1994, 12, 991–1045. [Google Scholar] [CrossRef]
- Jarrossay, D.; Napolitani, G.; Colonna, M.; Sallusto, F.; Lanzavecchia, A. Specialization and Complementarity in Microbial Molecule Recognition by Human Myeloid and Plasmacytoid Dendritic Cells. Eur. J. Immunol. 2001, 31, 3388–3393. [Google Scholar] [CrossRef]
- Kadowaki, N.; Antonenko, S.; Lau, J.Y.; Liu, Y.J. Natural Interferon Alpha/Beta-Producing Cells Link Innate and Adaptive Immunity. J. Exp. Med. 2000, 192, 219–226. [Google Scholar] [CrossRef]
- Gilliet, M.; Cao, W.; Liu, Y.-J. Plasmacytoid Dendritic Cells: Sensing Nucleic Acids in Viral Infection and Autoimmune Diseases. Nat. Rev. Immunol. 2008, 8, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, Z.J. The CGAS-CGAMP-STING Pathway Connects DNA Damage to Inflammation, Senescence, and Cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef] [PubMed]
- Komada, T.; Chung, H.; Lau, A.; Platnich, J.M.; Beck, P.L.; Benediktsson, H.; Duff, H.J.; Jenne, C.N.; Muruve, D.A. Macrophage Uptake of Necrotic Cell DNA Activates the AIM2 Inflammasome to Regulate a Proinflammatory Phenotype in CKD. J. Am. Soc. Nephrol. 2018, 29, 1165–1181. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.-R.; Fuertes, M.B.; Corrales, L.; Spranger, S.; Furdyna, M.J.; Leung, M.Y.K.; Duggan, R.; Wang, Y.; Barber, G.N.; Fitzgerald, K.A.; et al. STING-Dependent Cytosolic DNA Sensing Mediates Innate Immune Recognition of Immunogenic Tumors. Immunity 2014, 41, 830–842. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Cheng, H.; Yuan, H.; Xu, Q.; Shu, C.; Zhang, Y.; Xu, P.; Tan, J.; Rui, Y.; Li, P.; et al. Antitumor Activity of CGAMP via Stimulation of CGAS-CGAMP-STING-IRF3 Mediated Innate Immune Response. Sci. Rep. 2016, 6, 19049. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.; Xia, T.; Konno, H.; Konno, K.; Ruiz, P.; Barber, G.N. Inflammation-Driven Carcinogenesis Is Mediated through STING. Nat. Commun. 2014, 5, 5166. [Google Scholar] [CrossRef] [Green Version]
- Larkin, B.; Ilyukha, V.; Sorokin, M.; Buzdin, A.; Vannier, E.; Poltorak, A. Cutting Edge: Activation of STING in T Cells Induces Type I IFN Responses and Cell Death. J. Immunol. 2017, 199, 397–402. [Google Scholar] [CrossRef] [Green Version]
- Villani, A.-C.; Satija, R.; Reynolds, G.; Sarkizova, S.; Shekhar, K.; Fletcher, J.; Griesbeck, M.; Butler, A.; Zheng, S.; Lazo, S.; et al. Single-Cell RNA-Seq Reveals New Types of Human Blood Dendritic Cells, Monocytes, and Progenitors. Science 2017, 356, eaah4573. [Google Scholar] [CrossRef] [Green Version]
- Alcántara-Hernández, M.; Leylek, R.; Wagar, L.E.; Engleman, E.G.; Keler, T.; Marinkovich, M.P.; Davis, M.M.; Nolan, G.P.; Idoyaga, J. High-Dimensional Phenotypic Mapping of Human Dendritic Cells Reveals Interindividual Variation and Tissue Specialization. Immunity 2017, 47, 1037–1050.e6. [Google Scholar] [CrossRef] [Green Version]
- Leylek, R.; Alcántara-Hernández, M.; Lanzar, Z.; Lüdtke, A.; Perez, O.A.; Reizis, B.; Idoyaga, J. Integrated Cross-Species Analysis Identifies a Conserved Transitional Dendritic Cell Population. Cell Rep. 2019, 29, 3736–3750.e8. [Google Scholar] [CrossRef] [Green Version]
- Garnache-Ottou, F.; Vidal, C.; Biichlé, S.; Renosi, F.; Poret, E.; Pagadoy, M.; Desmarets, M.; Roggy, A.; Seilles, E.; Soret, L.; et al. How Should We Diagnose and Treat Blastic Plasmacytoid Dendritic Cell Neoplasm Patients? Blood Adv. 2019, 3, 4238–4251. [Google Scholar] [CrossRef] [Green Version]
- Renosi, F.; Roggy, A.; Giguelay, A.; Soret, L.; Viailly, P.-J.; Cheok, M.; Biichle, S.; Angelot-Delettre, F.; Asnafi, V.; Macintyre, E.; et al. Transcriptomic and Genomic Heterogeneity in Blastic Plasmacytoid Dendritic Cell Neoplasms: From Ontogeny to Oncogenesis. Blood Adv. 2021, 5, 1540–1551. [Google Scholar] [CrossRef]
- Sapienza, M.R.; Pileri, A.; Derenzini, E.; Melle, F.; Motta, G.; Fiori, S.; Calleri, A.; Pimpinelli, N.; Tabanelli, V.; Pileri, S. Blastic Plasmacytoid Dendritic Cell Neoplasm: State of the Art and Prospects. Cancers 2019, 11, 595. [Google Scholar] [CrossRef] [Green Version]
- Ceribelli, M.; Hou, Z.E.; Kelly, P.N.; Huang, D.W.; Wright, G.; Ganapathi, K.; Evbuomwan, M.O.; Pittaluga, S.; Shaffer, A.L.; Marcucci, G.; et al. A Druggable TCF4- and BRD4-Dependent Transcriptional Network Sustains Malignancy in Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancer Cell 2016, 30, 764–778. [Google Scholar] [CrossRef] [Green Version]
- Cota, C.; Vale, E.; Viana, I.; Requena, L.; Ferrara, G.; Anemona, L.; Metze, D.; Fink-Puches, R.; Wiesner, T.; Cerroni, L. Cutaneous Manifestations of Blastic Plasmacytoid Dendritic Cell Neoplasm-Morphologic and Phenotypic Variability in a Series of 33 Patients. Am. J. Surg. Pathol. 2010, 34, 75–87. [Google Scholar] [CrossRef]
- Dijkman, R.; van Doorn, R.; Szuhai, K.; Willemze, R.; Vermeer, M.H.; Tensen, C.P. Gene-Expression Profiling and Array-Based CGH Classify CD4+CD56+ Hematodermic Neoplasm and Cutaneous Myelomonocytic Leukemia as Distinct Disease Entities. Blood 2007, 109, 1720–1727. [Google Scholar] [CrossRef] [Green Version]
- Sapienza, M.R.; Fuligni, F.; Agostinelli, C.; Tripodo, C.; Righi, S.; Laginestra, M.A.; Pileri, A.; Mancini, M.; Rossi, M.; Ricci, F.; et al. Molecular Profiling of Blastic Plasmacytoid Dendritic Cell Neoplasm Reveals a Unique Pattern and Suggests Selective Sensitivity to NF-KB Pathway Inhibition. Leukemia 2014, 28, 1606–1616. [Google Scholar] [CrossRef] [Green Version]
- Philippe, L.; Ceroi, A.; Bôle-Richard, E.; Jenvrin, A.; Biichle, S.; Perrin, S.; Limat, S.; Bonnefoy, F.; Deconinck, E.; Saas, P.; et al. Bortezomib as a New Therapeutic Approach for Blastic Plasmacytoid Dendritic Cell Neoplasm. Haematologica 2017, 102, 1861–1868. [Google Scholar] [CrossRef] [Green Version]
- Montero, J.; Stephansky, J.; Cai, T.; Griffin, G.K.; Cabal-Hierro, L.; Togami, K.; Hogdal, L.J.; Galinsky, I.; Morgan, E.A.; Aster, J.C.; et al. Blastic Plasmacytoid Dendritic Cell Neoplasm Is Dependent on BCL2 and Sensitive to Venetoclax. Cancer Discov. 2017, 7, 156–164. [Google Scholar] [CrossRef] [Green Version]
- Pemmaraju, N.; Lane, A.A.; Sweet, K.L.; Stein, A.S.; Vasu, S.; Blum, W.; Rizzieri, D.A.; Wang, E.S.; Duvic, M.; Sloan, J.M.; et al. Tagraxofusp in Blastic Plasmacytoid Dendritic-Cell Neoplasm. N. Engl. J. Med. 2019, 380, 1628–1637. [Google Scholar] [CrossRef]
- Bôle-Richard, E.; Pemmaraju, N.; Caël, B.; Daguindau, E.; Lane, A.A. CD123 and More: How to Target the Cell Surface of Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancers 2022, 14, 2287. [Google Scholar] [CrossRef]
- Zhou, B.; Lawrence, T.; Liang, Y. The Role of Plasmacytoid Dendritic Cells in Cancers. Front. Immunol. 2021, 12, 749190. [Google Scholar] [CrossRef]
- Faget, J.; Sisirak, V.; Blay, J.-Y.; Caux, C.; Bendriss-Vermare, N.; Ménétrier-Caux, C. ICOS Is Associated with Poor Prognosis in Breast Cancer as It Promotes the Amplification of Immunosuppressive CD4+ T Cells by Plasmacytoid Dendritic Cells. Oncoimmunology 2013, 2, e23185. [Google Scholar] [CrossRef] [Green Version]
- Labidi-Galy, S.I.; Treilleux, I.; Goddard-Leon, S.; Combes, J.-D.; Blay, J.-Y.; Ray-Coquard, I.; Caux, C.; Bendriss-Vermare, N. Plasmacytoid Dendritic Cells Infiltrating Ovarian Cancer Are Associated with Poor Prognosis. Oncoimmunology 2012, 1, 380–382. [Google Scholar] [CrossRef] [Green Version]
- Gerlini, G.; Urso, C.; Mariotti, G.; Di Gennaro, P.; Palli, D.; Brandani, P.; Salvadori, A.; Pimpinelli, N.; Reali, U.M.; Borgognoni, L. Plasmacytoid Dendritic Cells Represent a Major Dendritic Cell Subset in Sentinel Lymph Nodes of Melanoma Patients and Accumulate in Metastatic Nodes. Clin. Immunol. 2007, 125, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Guéry, L.; Dubrot, J.; Lippens, C.; Brighouse, D.; Malinge, P.; Irla, M.; Pot, C.; Reith, W.; Waldburger, J.-M.; Hugues, S. Ag-Presenting CpG-Activated PDCs Prime Th17 Cells That Induce Tumor Regression. Cancer Res. 2014, 74, 6430–6440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poropatich, K.; Dominguez, D.; Chan, W.-C.; Andrade, J.; Zha, Y.; Wray, B.; Miska, J.; Qin, L.; Cole, L.; Coates, S.; et al. OX40+ Plasmacytoid Dendritic Cells in the Tumor Microenvironment Promote Antitumor Immunity. J. Clin. Investig. 2020, 130, 3528–3542. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-C. The Non-Canonical NF-ΚB Pathway in Immunity and Inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Katakam, A.K.; Brightbill, H.; Franci, C.; Kung, C.; Nunez, V.; Jones, C.; Peng, I.; Jeet, S.; Wu, L.C.; Mellman, I.; et al. Dendritic Cells Require NIK for CD40-Dependent Cross-Priming of CD8+ T Cells. Proc. Natl. Acad. Sci. USA 2015, 112, 14664–14669. [Google Scholar] [CrossRef] [Green Version]
- Fu, C.; Peng, P.; Loschko, J.; Feng, L.; Pham, P.; Cui, W.; Lee, K.P.; Krug, A.B.; Jiang, A. Plasmacytoid Dendritic Cells Cross-Prime Naive CD8 T Cells by Transferring Antigen to Conventional Dendritic Cells through Exosomes. Proc. Natl. Acad. Sci. USA 2020, 117, 23730–23741. [Google Scholar] [CrossRef]
- Ito, T.; Yang, M.; Wang, Y.-H.; Lande, R.; Gregorio, J.; Perng, O.A.; Qin, X.-F.; Liu, Y.-J.; Gilliet, M. Plasmacytoid Dendritic Cells Prime IL-10-Producing T Regulatory Cells by Inducible Costimulator Ligand. J. Exp. Med. 2007, 204, 105–115. [Google Scholar] [CrossRef]
- Munn, D.H.; Sharma, M.D.; Hou, D.; Baban, B.; Lee, J.R.; Antonia, S.J.; Messina, J.L.; Chandler, P.; Koni, P.A.; Mellor, A.L. Expression of Indoleamine 2,3-Dioxygenase by Plasmacytoid Dendritic Cells in Tumor-Draining Lymph Nodes. J. Clin. Investig. 2004, 114, 280–290. [Google Scholar] [CrossRef] [Green Version]
- Conrad, C.; Gregorio, J.; Wang, Y.-H.; Ito, T.; Meller, S.; Hanabuchi, S.; Anderson, S.; Atkinson, N.; Ramirez, P.T.; Liu, Y.-J.; et al. Plasmacytoid Dendritic Cells Promote Immunosuppression in Ovarian Cancer via ICOS Costimulation of Foxp3(+) T-Regulatory Cells. Cancer Res. 2012, 72, 5240–5249. [Google Scholar] [CrossRef] [Green Version]
- Nagase, H.; Takeoka, T.; Urakawa, S.; Morimoto-Okazawa, A.; Kawashima, A.; Iwahori, K.; Takiguchi, S.; Nishikawa, H.; Sato, E.; Sakaguchi, S.; et al. ICOS+ Foxp3+ TILs in Gastric Cancer Are Prognostic Markers and Effector Regulatory T Cells Associated with Helicobacter Pylori. Int. J. Cancer 2017, 140, 686–695. [Google Scholar] [CrossRef]
- Baban, B.; Chandler, P.R.; Sharma, M.D.; Pihkala, J.; Koni, P.A.; Munn, D.H.; Mellor, A.L. IDO Activates Regulatory T Cells and Blocks Their Conversion into Th17-like T Cells. J. Immunol. 2009, 183, 2475–2483. [Google Scholar] [CrossRef] [Green Version]
- Meireson, A.; Devos, M.; Brochez, L. IDO Expression in Cancer: Different Compartment, Different Functionality? Front. Immunol. 2020, 11, 531491. [Google Scholar] [CrossRef]
- Sisirak, V.; Vey, N.; Goutagny, N.; Renaudineau, S.; Malfroy, M.; Thys, S.; Treilleux, I.; Labidi-Galy, S.I.; Bachelot, T.; Dezutter-Dambuyant, C.; et al. Breast Cancer-Derived Transforming Growth Factor-β and Tumor Necrosis Factor-α Compromise Interferon-α Production by Tumor-Associated Plasmacytoid Dendritic Cells. Int. J. Cancer 2013, 133, 771–778. [Google Scholar] [CrossRef] [Green Version]
- Durham, N.M.; Holoweckyj, N.; MacGill, R.S.; McGlinchey, K.; Leow, C.C.; Robbins, S.H. GITR Ligand Fusion Protein Agonist Enhances the Tumor Antigen–Specific CD8 T-Cell Response and Leads to Long-Lasting Memory. J. Immunother. Cancer 2017, 5, 47. [Google Scholar] [CrossRef]
- Heeren, A.M.; Rotman, J.; Samuels, S.; Zijlmans, H.J.M.A.A.; Fons, G.; van de Vijver, K.K.; Bleeker, M.C.G.; Kenter, G.G.; Jordanova, E.J.; de Gruijl, T.D. Immune Landscape in Vulvar Cancer-Draining Lymph Nodes Indicates Distinct Immune Escape Mechanisms in Support of Metastatic Spread and Growth. J. Immunother. Cancer 2021, 9, e003623. [Google Scholar] [CrossRef]
- Wei, S.; Kryczek, I.; Zou, L.; Daniel, B.; Cheng, P.; Mottram, P.; Curiel, T.; Lange, A.; Zou, W. Plasmacytoid Dendritic Cells Induce CD8+ Regulatory T Cells in Human Ovarian Carcinoma. Cancer Res. 2005, 65, 5020–5026. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Ke, X.; Zeng, S.; Wu, M.; Lou, J.; Wu, L.; Huang, P.; Huang, L.; Wang, F.; Pan, S. Analysis of CD8+ Treg Cells in Patients with Ovarian Cancer: A Possible Mechanism for Immune Impairment. Cell. Mol. Immunol. 2015, 12, 580–591. [Google Scholar] [CrossRef] [Green Version]
- Barilla, R.M.; Diskin, B.; Caso, R.C.; Lee, K.B.; Mohan, N.; Buttar, C.; Adam, S.; Sekendiz, Z.; Wang, J.; Salas, R.D.; et al. Specialized Dendritic Cells Induce Tumor-Promoting IL-10+IL-17+ FoxP3neg Regulatory CD4+ T Cells in Pancreatic Carcinoma. Nat. Commun. 2019, 10, 1424. [Google Scholar] [CrossRef] [Green Version]
- Van Gulijk, M.; Dammeijer, F.; Aerts, J.G.J.V.; Vroman, H. Combination Strategies to Optimize Efficacy of Dendritic Cell-Based Immunotherapy. Front. Immunol. 2018, 9, 2759. [Google Scholar] [CrossRef] [Green Version]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic Cells in Cancer Immunology and Immunotherapy. Nat. Rev. Immunol. 2019, 20, 7–24. [Google Scholar] [CrossRef]
- Summers deLuca, L.; Gommerman, J.L. Fine-Tuning of Dendritic Cell Biology by the TNF Superfamily. Nat. Rev. Immunol. 2012, 12, 339–351. [Google Scholar] [CrossRef]
- Bol, K.F.; Schreibelt, G.; Gerritsen, W.R.; de Vries, I.J.M.; Figdor, C.G. Dendritic Cell-Based Immunotherapy: State of the Art and Beyond. Clin. Cancer Res. 2016, 22, 1897–1906. [Google Scholar] [CrossRef] [Green Version]
- Block, M.S.; Dietz, A.B.; Gustafson, M.P.; Kalli, K.R.; Erskine, C.L.; Youssef, B.; Vijay, G.V.; Allred, J.B.; Pavelko, K.D.; Strausbauch, M.A.; et al. Th17-Inducing Autologous Dendritic Cell Vaccination Promotes Antigen-Specific Cellular and Humoral Immunity in Ovarian Cancer Patients. Nat. Commun. 2020, 11, 5173. [Google Scholar] [CrossRef]
- Brincks, E.L.; Adams, J.; Wang, L.; Turner, B.; Marcinowicz, A.; Ke, J.; Essmann, M.; Mautino, L.M.; Allen, C.V.; Kumar, S.; et al. Indoximod Opposes the Immunosuppressive Effects Mediated by IDO and TDO via Modulation of AhR Function and Activation of MTORC1. Oncotarget 2020, 11, 2438–2461. [Google Scholar] [CrossRef]
- Grobben, Y.; de Man, J.; van Doornmalen, A.M.; Muller, M.; Willemsen-Seegers, N.; Vu-Pham, D.; Mulder, W.R.; Prinsen, M.B.W.; de Wit, J.; Sterrenburg, J.G.; et al. Targeting Indoleamine 2,3-Dioxygenase in Cancer Models Using the Novel Small Molecule Inhibitor NTRC 3883-0. Front. Immunol. 2020, 11, 609490. [Google Scholar] [CrossRef]
- Zakharia, Y.; McWilliams, R.R.; Rixe, O.; Drabick, J.; Shaheen, M.F.; Grossmann, K.F.; Kolhe, R.; Pacholczyk, R.; Sadek, R.; Tennant, L.L.; et al. Phase II Trial of the IDO Pathway Inhibitor Indoximod plus Pembrolizumab for the Treatment of Patients with Advanced Melanoma. J. Immunother. Cancer 2021, 9, e002057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, M.; Wang, F. Immune Regulation by CD8+ Treg Cells: Novel Possibilities for Anticancer Immunotherapy. Cell. Mol. Immunol. 2018, 15, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Lucas, N.; Duchmann, M.; Rameau, P.; Noël, F.; Michea, P.; Saada, V.; Kosmider, O.; Pierron, G.; Fernandez-Zapico, M.E.; Howard, M.T.; et al. Biology and Prognostic Impact of Clonal Plasmacytoid Dendritic Cells in Chronic Myelomonocytic Leukemia. Leukemia 2019, 33, 2466–2480. [Google Scholar] [CrossRef]
- Chan, A.; Liu, Y.; Devlin, S.; Gao, Q.; Baik, J.; Sigler, A.; Londono, D.; Arcila, M.; Levine, R.; Dogan, A.; et al. Reduced Plasmacytoid Dendritic Cell Output Is Associated With High Risk in Low-Grade Myelodysplastic Syndrome. HemaSphere 2022, 6, e685. [Google Scholar] [CrossRef] [PubMed]
- Hamadeh, F.; Awadallah, A.; Meyerson, H.J.; Beck, R.C. Flow Cytometry Identifies a Spectrum of Maturation in Myeloid Neoplasms Having Plasmacytoid Dendritic Cell Differentiation. Cytom. B Clin. Cytom. 2020, 98, 43–51. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Y.; Chang, Y.; Yuan, X.; Hao, L.; Shi, H.; Lai, Y.; Huang, X.; Liu, Y. Myeloid Neoplasms with Elevated Plasmacytoid Dendritic Cell Differentiation Reflect the Maturation Process of Dendritic Cells. Cytom. A 2020, 97, 61–69. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Tefferi, A. Chronic Myelomonocytic Leukemia: 2022 Update on Diagnosis, Risk Stratification, and Management. Am. J. Hematol. 2022, 97, 352–372. [Google Scholar] [CrossRef]
- Elena, C.; Gallì, A.; Such, E.; Meggendorfer, M.; Germing, U.; Rizzo, E.; Cervera, J.; Molteni, E.; Fasan, A.; Schuler, E.; et al. Integrating Clinical Features and Genetic Lesions in the Risk Assessment of Patients with Chronic Myelomonocytic Leukemia. Blood 2016, 128, 1408–1417. [Google Scholar] [CrossRef]
- Chan, O.; Renneville, A.; Padron, E. Chronic Myelomonocytic Leukemia Diagnosis and Management. Leukemia 2021, 35, 1552–1562. [Google Scholar] [CrossRef]
- Yang, H.; Bueso-Ramos, C.; DiNardo, C.; Estecio, M.R.; Davanlou, M.; Geng, Q.-R.; Fang, Z.; Nguyen, M.; Pierce, S.; Wei, Y.; et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in Myelodysplastic Syndromes Is Enhanced by Treatment with Hypomethylating Agents. Leukemia 2014, 28, 1280–1288. [Google Scholar] [CrossRef]
- Droin, N.; Jacquel, A.; Hendra, J.-B.; Racoeur, C.; Truntzer, C.; Pecqueur, D.; Benikhlef, N.; Ciudad, M.; Guery, L.; Jooste, V.; et al. Alpha-Defensins Secreted by Dysplastic Granulocytes Inhibit the Differentiation of Monocytes in Chronic Myelomonocytic Leukemia. Blood 2010, 115, 78–88. [Google Scholar] [CrossRef] [Green Version]
- Vuckovic, S.; Fearnley, D.B.; Gunningham, S.; Spearing, R.L.; Patton, W.N.; Hart, D.N. Dendritic Cells in Chronic Myelomonocytic Leukaemia. Br. J. Haematol. 1999, 105, 974–985. [Google Scholar] [CrossRef]
- Orazi, A.; Chiu, R.; O’Malley, D.P.; Czader, M.; Allen, S.L.; An, C.; Vance, G.H. Chronic Myelomonocytic Leukemia: The Role of Bone Marrow Biopsy Immunohistology. Mod. Pathol. 2006, 19, 1536–1545. [Google Scholar] [CrossRef]
- Mangaonkar, A.A.; Patnaik, M.M. Advances in Chronic Myelomonocytic Leukemia and Future Prospects: Lessons Learned from Precision Genomics. Adv. Cell Gene Ther. 2019, 2, e48. [Google Scholar] [CrossRef]
- Mangaonkar, A.A.; Reichard, K.K.; Binder, M.; Coltro, G.; Lasho, T.L.; Carr, R.M.; Chiu, A.; Negron, V.; Hefazi, M.; Anagnostou, T.; et al. Bone Marrow Dendritic Cell Aggregates Associate with Systemic Immune Dysregulation in Chronic Myelomonocytic Leukemia. Blood Adv. 2020, 4, 5425–5430. [Google Scholar] [CrossRef]
- You, X.; Liu, F.; Binder, M.; Vedder, A.; Lasho, T.; Wen, Z.; Gao, X.; Flietner, E.; Rajagopalan, A.; Zhou, Y.; et al. Asxl1 Loss Cooperates with Oncogenic Nras in Mice to Reprogram the Immune Microenvironment and Drive Leukemic Transformation. Blood 2022, 139, 1066–1079. [Google Scholar] [CrossRef] [PubMed]
- Mangaonkar, A.A.; Patnaik, M.M. Role of the Bone Marrow Immune Microenvironment in Chronic Myelomonocytic Leukemia Pathogenesis: Novel Mechanisms and Insights into Clonal Propagation. Leuk. Lymphoma 2022, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Beau, M.M.L.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Gañán-Gómez, I.; Wei, Y.; Starczynowski, D.T.; Colla, S.; Yang, H.; Cabrero-Calvo, M.; Bohannan, Z.S.; Verma, A.; Steidl, U.; Garcia-Manero, G. Deregulation of Innate Immune and Inflammatory Signaling in Myelodysplastic Syndromes. Leukemia 2015, 29, 1458–1469. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Estey, E.; Wen, S.; Pierce, S.; Kantarjian, H.; Albitar, M.; Kurzrock, R. The Prognostic Significance of Cytokine Levels in Newly Diagnosed Acute Myeloid Leukemia and High-Risk Myelodysplastic Syndromes. Cancer 2008, 113, 1605–1613. [Google Scholar] [CrossRef]
- Shetty, V.; Mundle, S.; Alvi, S.; Showel, M.; Broady-Robinson, L.; Dar, S.; Borok, R.; Showel, J.; Gregory, S.; Rifkin, S.; et al. Measurement of Apoptosis, Proliferation and Three Cytokines in 46 Patients with Myelodysplastic Syndromes. Leuk. Res. 1996, 20, 891–900. [Google Scholar] [CrossRef]
- Kornblau, S.M.; McCue, D.; Singh, N.; Chen, W.; Estrov, Z.; Coombes, K.R. Recurrent Expression Signatures of Cytokines and Chemokines Are Present and Are Independently Prognostic in Acute Myelogenous Leukemia and Myelodysplasia. Blood 2010, 116, 4251–4261. [Google Scholar] [CrossRef] [Green Version]
- Kordasti, S.Y.; Afzali, B.; Lim, Z.; Ingram, W.; Hayden, J.; Barber, L.; Matthews, K.; Chelliah, R.; Guinn, B.; Lombardi, G.; et al. IL-17-Producing CD4(+) T Cells, pro-Inflammatory Cytokines and Apoptosis Are Increased in Low Risk Myelodysplastic Syndrome. Br. J. Haematol. 2009, 145, 64–72. [Google Scholar] [CrossRef]
- Berthon, C.; Fontenay, M.; Corm, S.; Briche, I.; Allorge, D.; Hennart, B.; Lhermitte, M.; Quesnel, B. Metabolites of Tryptophan Catabolism Are Elevated in Sera of Patients with Myelodysplastic Syndromes and Inhibit Hematopoietic Progenitor Amplification. Leuk. Res. 2013, 37, 573–579. [Google Scholar] [CrossRef]
- Maratheftis, C.I.; Andreakos, E.; Moutsopoulos, H.M.; Voulgarelis, M. Toll-like Receptor-4 Is up-Regulated in Hematopoietic Progenitor Cells and Contributes to Increased Apoptosis in Myelodysplastic Syndromes. Clin. Cancer Res. 2007, 13, 1154–1160. [Google Scholar] [CrossRef] [Green Version]
- Kuninaka, N.; Kurata, M.; Yamamoto, K.; Suzuki, S.; Umeda, S.; Kirimura, S.; Arai, A.; Nakagawa, Y.; Suzuki, K.; Kitagawa, M. Expression of Toll-like Receptor 9 in Bone Marrow Cells of Myelodysplastic Syndromes Is down-Regulated during Transformation to Overt Leukemia. Exp. Mol. Pathol. 2010, 88, 293–298. [Google Scholar] [CrossRef]
- Sioud, M.; Fløisand, Y.; Forfang, L.; Lund-Johansen, F. Signaling through Toll-like Receptor 7/8 Induces the Differentiation of Human Bone Marrow CD34+ Progenitor Cells along the Myeloid Lineage. J. Mol. Biol. 2006, 364, 945–954. [Google Scholar] [CrossRef]
- Stein, S.J.; Baldwin, A.S. Deletion of the NF-ΚB Subunit P65/RelA in the Hematopoietic Compartment Leads to Defects in Hematopoietic Stem Cell Function. Blood 2013, 121, 5015–5024. [Google Scholar] [CrossRef] [Green Version]
- Trowbridge, J.J.; Starczynowski, D.T. Innate Immune Pathways and Inflammation in Hematopoietic Aging, Clonal Hematopoiesis, and MDS. J. Exp. Med. 2021, 218, e20201544. [Google Scholar] [CrossRef]
- Ma, L.; Delforge, M.; van Duppen, V.; Verhoef, G.; Emanuel, B.; Boogaerts, M.; Hagemeijer, A.; Vandenberghe, P. Circulating Myeloid and Lymphoid Precursor Dendritic Cells Are Clonally Involved in Myelodysplastic Syndromes. Leukemia 2004, 18, 1451–1456. [Google Scholar] [CrossRef] [Green Version]
- Saft, L.; Björklund, E.; Berg, E.; Hellström-Lindberg, E.; Porwit, A. Bone Marrow Dendritic Cells Are Reduced in Patients with High-Risk Myelodysplastic Syndromes. Leuk. Res. 2013, 37, 266–273. [Google Scholar] [CrossRef]
- Carenza, C.; Calcaterra, F.; Oriolo, F.; Di Vito, C.; Ubezio, M.; Della Porta, M.G.; Mavilio, D.; Della Bella, S. Costimulatory Molecules and Immune Checkpoints Are Differentially Expressed on Different Subsets of Dendritic Cells. Front. Immunol. 2019, 10, 1325. [Google Scholar] [CrossRef]
- Van Leeuwen-Kerkhoff, N.; Westers, T.M.; Poddighe, P.J.; Povoleri, G.A.M.; Timms, J.A.; Kordasti, S.; de Gruijl, T.D.; van de Loosdrecht, A.A. Reduced Frequencies and Functional Impairment of Dendritic Cell Subsets and Non-Classical Monocytes in Myelodysplastic Syndromes. Haematologica 2022, 107, 655–667. [Google Scholar] [CrossRef]
- Agarwal, A.; Bolosky, W.J.; Wilson, D.B.; Eide, C.A.; Olson, S.B.; Fan, G.; Druker, B.J. Differentiation of Leukemic Blasts Is Not Completely Blocked in Acute Myeloid Leukemia. Proc. Natl. Acad. Sci. USA 2019, 116, 24593–24599. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [Green Version]
- Roussel, X.; Daguindau, E.; Berceanu, A.; Desbrosses, Y.; Warda, W.; Neto da Rocha, M.; Trad, R.; Deconinck, E.; Deschamps, M.; Ferrand, C. Acute Myeloid Leukemia: From Biology to Clinical Practices Through Development and Pre-Clinical Therapeutics. Front. Oncol. 2020, 10, 599933. [Google Scholar] [CrossRef]
- Martín-Martín, L.; Almeida, J.; Hernández-Campo, P.M.; Sánchez, M.L.; Lécrevisse, Q.; Orfao, A. Immunophenotypical, Morphologic, and Functional Characterization of Maturation-Associated Plasmacytoid Dendritic Cell Subsets in Normal Adult Human Bone Marrow. Transfusion 2009, 49, 1692–1708. [Google Scholar] [CrossRef]
- Waidhauser, J.; Labopin, M.; Esteve, J.; Kröger, N.; Cornelissen, J.; Gedde-Dahl, T.; Van Gorkom, G.; Finke, J.; Rovira, M.; Schaap, N.; et al. Allogeneic Stem Cell Transplantation for AML Patients with RUNX1 Mutation in First Complete Remission: A Study on Behalf of the Acute Leukemia Working Party of the EBMT. Bone Marrow Transpl. 2021, 56, 2445–2453. [Google Scholar] [CrossRef]
- Hoelzer, D.; Bassan, R.; Dombret, H.; Fielding, A.; Ribera, J.M.; Buske, C. Acute Lymphoblastic Leukaemia in Adult Patients: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-up. Ann. Oncol. 2016, 27, v69–v82. [Google Scholar] [CrossRef]
- Michelozzi, I.M.; Granata, V.; De Ponti, G.; Alberti, G.; Tomasoni, C.; Antolini, L.; Gambacorti-Passerini, C.; Gentner, B.; Dazzi, F.; Biondi, A.; et al. Acute Myeloid Leukaemia Niche Regulates Response to L-Asparaginase. Br. J. Haematol. 2019, 186, 420–430. [Google Scholar] [CrossRef]
- Bôle-Richard, E.; Fredon, M.; Biichlé, S.; Anna, F.; Certoux, J.-M.; Renosi, F.; Tsé, F.; Molimard, C.; Valmary-Degano, S.; Jenvrin, A.; et al. CD28/4-1BB CD123 CAR T Cells in Blastic Plasmacytoid Dendritic Cell Neoplasm. Leukemia 2020, 34, 3228–3241. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G. CD123 as a Therapeutic Target in the Treatment of Hematological Malignancies. Cancers 2019, 11, 1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brauchle, B.; Goldstein, R.L.; Karbowski, C.M.; Henn, A.; Li, C.-M.; Bücklein, V.L.; Krupka, C.; Boyle, M.C.; Koppikar, P.; Haubner, S.; et al. Characterization of a Novel FLT3 BiTE Molecule for the Treatment of Acute Myeloid Leukemia. Mol. Cancer Ther. 2020, 19, 1875–1888. [Google Scholar] [CrossRef]
- Curti, A.; Trabanelli, S.; Onofri, C.; Aluigi, M.; Salvestrini, V.; Ocadlikova, D.; Evangelisti, C.; Rutella, S.; De Cristofaro, R.; Ottaviani, E.; et al. Indoleamine 2,3-Dioxygenase-Expressing Leukemic Dendritic Cells Impair a Leukemia-Specific Immune Response by Inducing Potent T Regulatory Cells. Haematologica 2010, 95, 2022–2030. [Google Scholar] [CrossRef] [Green Version]
- Wells, G.; Kennedy, P.T.; Dahal, L.N. Investigating the Role of Indoleamine 2,3-Dioxygenase in Acute Myeloid Leukemia: A Systematic Review. Front. Immunol. 2021, 12, 651687. [Google Scholar] [CrossRef]
- Williams, P.; Basu, S.; Garcia-Manero, G.; Hourigan, C.S.; Oetjen, K.A.; Cortes, J.E.; Ravandi, F.; Jabbour, E.J.; Al-Hamal, Z.; Konopleva, M.; et al. The Distribution of T-Cell Subsets and the Expression of Immune Checkpoint Receptors and Ligands in Patients with Newly Diagnosed and Relapsed Acute Myeloid Leukemia. Cancer 2019, 125, 1470–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Dong, Y.; Yang, Q.; Xu, W.; Jiang, S.; Yu, Z.; Yu, K.; Zhang, S. Acute Myeloid Leukemia Cells Express ICOS Ligand to Promote the Expansion of Regulatory T Cells. Front. Immunol. 2018, 9, 2227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Jia, B.; Claxton, D.F.; Ehmann, W.C.; Rybka, W.B.; Mineishi, S.; Naik, S.; Khawaja, M.R.; Sivik, J.; Han, J.; et al. VISTA Is Highly Expressed on MDSCs and Mediates an Inhibition of T Cell Response in Patients with AML. Oncoimmunology 2018, 7, e1469594. [Google Scholar] [CrossRef] [PubMed]
- Brauneck, F.; Haag, F.; Woost, R.; Wildner, N.; Tolosa, E.; Rissiek, A.; Vohwinkel, G.; Wellbrock, J.; Bokemeyer, C.; Schulze zur Wiesch, J.; et al. Increased Frequency of TIGIT+CD73-CD8+ T Cells with a TOX+ TCF-1low Profile in Patients with Newly Diagnosed and Relapsed AML. Oncoimmunology 2021, 10, 1930391. [Google Scholar] [CrossRef]
- Chao, M.P.; Takimoto, C.H.; Feng, D.D.; McKenna, K.; Gip, P.; Liu, J.; Volkmer, J.-P.; Weissman, I.L.; Majeti, R. Therapeutic Targeting of the Macrophage Immune Checkpoint CD47 in Myeloid Malignancies. Front. Oncol. 2019, 9, 1380. [Google Scholar] [CrossRef]
- Vago, L.; Gojo, I. Immune Escape and Immunotherapy of Acute Myeloid Leukemia. J. Clin. Investig. 2020, 130, 1552–1564. [Google Scholar] [CrossRef]
- Daver, N.; Alotaibi, A.S.; Bücklein, V.; Subklewe, M. T-Cell-Based Immunotherapy of Acute Myeloid Leukemia: Current Concepts and Future Developments. Leukemia 2021, 35, 1843–1863. [Google Scholar] [CrossRef]
- Abbas, H.A.; Hao, D.; Tomczak, K.; Barrodia, P.; Im, J.S.; Reville, P.K.; Alaniz, Z.; Wang, W.; Wang, R.; Wang, F.; et al. Single Cell T Cell Landscape and T Cell Receptor Repertoire Profiling of AML in Context of PD-1 Blockade Therapy. Nat. Commun. 2021, 12, 6071. [Google Scholar] [CrossRef]
- Stroopinsky, D.; Liegel, J.; Bhasin, M.; Cheloni, G.; Thomas, B.; Bhasin, S.; Panchal, R.; Ghiasuddin, H.; Rahimian, M.; Nahas, M.; et al. Leukemia Vaccine Overcomes Limitations of Checkpoint Blockade by Evoking Clonal T Cell Responses in a Murine Acute Myeloid Leukemia Model. Haematologica 2021, 106, 1330–1342. [Google Scholar] [CrossRef]
- Ehx, G.; Larouche, J.-D.; Durette, C.; Laverdure, J.-P.; Hesnard, L.; Vincent, K.; Hardy, M.-P.; Thériault, C.; Rulleau, C.; Lanoix, J.; et al. Atypical Acute Myeloid Leukemia-Specific Transcripts Generate Shared and Immunogenic MHC Class-I-Associated Epitopes. Immunity 2021, 54, 737–752.e10. [Google Scholar] [CrossRef]
- Yarchoan, M.; Johnson, B.A.; Lutz, E.R.; Laheru, D.A.; Jaffee, E.M. Targeting Neoantigens to Augment Antitumour Immunity. Nat. Rev. Cancer 2017, 17, 209–222. [Google Scholar] [CrossRef]
- Manfredi, F.; Cianciotti, B.C.; Potenza, A.; Tassi, E.; Noviello, M.; Biondi, A.; Ciceri, F.; Bonini, C.; Ruggiero, E. TCR Redirected T Cells for Cancer Treatment: Achievements, Hurdles, and Goals. Front. Immunol. 2020, 11, 1689. [Google Scholar] [CrossRef]
- Janelle, V.; Rulleau, C.; Del Testa, S.; Carli, C.; Delisle, J.-S. T-Cell Immunotherapies Targeting Histocompatibility and Tumor Antigens in Hematological Malignancies. Front. Immunol. 2020, 11, 276. [Google Scholar] [CrossRef] [Green Version]
- Gaidzik, V.I.; Bullinger, L.; Schlenk, R.F.; Zimmermann, A.S.; Röck, J.; Paschka, P.; Corbacioglu, A.; Krauter, J.; Schlegelberger, B.; Ganser, A.; et al. RUNX1 Mutations in Acute Myeloid Leukemia: Results From a Comprehensive Genetic and Clinical Analysis From the AML Study Group. JCO 2011, 29, 1364–1372. [Google Scholar] [CrossRef]
- Gaidzik, V.I.; Teleanu, V.; Papaemmanuil, E.; Weber, D.; Paschka, P.; Hahn, J.; Wallrabenstein, T.; Kolbinger, B.; Köhne, C.H.; Horst, H.A.; et al. RUNX1 Mutations in Acute Myeloid Leukemia Are Associated with Distinct Clinico-Pathologic and Genetic Features. Leukemia 2016, 30, 2160–2168. [Google Scholar] [CrossRef]
- Yokota, A.; Huo, L.; Lan, F.; Wu, J.; Huang, G. The Clinical, Molecular, and Mechanistic Basis of RUNX1 Mutations Identified in Hematological Malignancies. Mol. Cells 2020, 43, 145–152. [Google Scholar] [CrossRef]
- Simon, L.; Lavallée, V.-P.; Bordeleau, M.-E.; Krosl, J.; Baccelli, I.; Boucher, G.; Lehnertz, B.; Chagraoui, J.; MacRae, T.; Ruel, R.; et al. Chemogenomic Landscape of RUNX1-Mutated AML Reveals Importance of RUNX1 Allele Dosage in Genetics and Glucocorticoid Sensitivity. Clin. Cancer Res. 2017, 23, 6969–6981. [Google Scholar] [CrossRef] [Green Version]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in Hematological Malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef] [Green Version]
- Schnittger, S.; Dicker, F.; Kern, W.; Wendland, N.; Sundermann, J.; Alpermann, T.; Haferlach, C.; Haferlach, T. RUNX1 Mutations Are Frequent in de Novo AML with Noncomplex Karyotype and Confer an Unfavorable Prognosis. Blood 2011, 117, 2348–2357. [Google Scholar] [CrossRef] [Green Version]
- Kamikubo, Y. Genetic Compensation of RUNX Family Transcription Factors in Leukemia. Cancer Sci. 2018, 109, 2358–2363. [Google Scholar] [CrossRef] [Green Version]
- Hass, M.R.; Brissette, D.; Parameswaran, S.; Pujato, M.; Donmez, O.; Kottyan, L.C.; Weirauch, M.T.; Kopan, R. Runx1 Shapes the Chromatin Landscape via a Cascade of Direct and Indirect Targets. PLOS Genet. 2021, 17, e1009574. [Google Scholar] [CrossRef]
- Hayashi, Y.; Harada, Y.; Harada, H. Myeloid Neoplasms and Clonal Hematopoiesis from the RUNX1 Perspective. Leukemia 2022, 36, 1203–1214. [Google Scholar] [CrossRef]
- Barreyro, L.; Chlon, T.M.; Starczynowski, D.T. Chronic Immune Response Dysregulation in MDS Pathogenesis. Blood 2018, 132, 1553–1560. [Google Scholar] [CrossRef] [Green Version]
- Montalban-Bravo, G.; Class, C.A.; Ganan-Gomez, I.; Kanagal-Shamanna, R.; Sasaki, K.; Richard-Carpentier, G.; Naqvi, K.; Wei, Y.; Yang, H.; Soltysiak, K.A.; et al. Transcriptomic Analysis Implicates Necroptosis in Disease Progression and Prognosis in Myelodysplastic Syndromes. Leukemia 2020, 34, 872–881. [Google Scholar] [CrossRef]
- Bellissimo, D.C.; Chen, C.-H.; Zhu, Q.; Bagga, S.; Lee, C.-T.; He, B.; Wertheim, G.B.; Jordan, M.; Tan, K.; Worthen, G.S.; et al. Runx1 Negatively Regulates Inflammatory Cytokine Production by Neutrophils in Response to Toll-like Receptor Signaling. Blood Adv. 2020, 4, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Kitoh, A.; Ono, M.; Naoe, Y.; Ohkura, N.; Yamaguchi, T.; Yaguchi, H.; Kitabayashi, I.; Tsukada, T.; Nomura, T.; Miyachi, Y.; et al. Indispensable Role of the Runx1-Cbfbeta Transcription Complex for in Vivo-Suppressive Function of FoxP3+ Regulatory T Cells. Immunity 2009, 31, 609–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, W.F.; Kohu, K.; Nakamura, A.; Ebina, M.; Kikuchi, T.; Tazawa, R.; Tanaka, K.; Kon, S.; Funaki, T.; Sugahara-Tobinai, A.; et al. Runx1 Deficiency in CD4+ T Cells Causes Fatal Autoimmune Inflammatory Lung Disease Due to Spontaneous Hyperactivation of Cells. J Immunol. 2012, 188, 5408–5420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Gao, L.; Teng, L.; Ge, J.; Oo, Z.M.; Kumar, A.R.; Gilliland, D.G.; Mason, P.J.; Tan, K.; Speck, N.A. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell 2015, 17, 165–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muto, T.; Walker, C.S.; Choi, K.; Hueneman, K.; Smith, M.A.; Gul, Z.; Garcia-Manero, G.; Ma, A.; Zheng, Y.; Starczynowski, D.T. Adaptive Response to Inflammation Contributes to Sustained Myelopoiesis and Confers a Competitive Advantage in Myelodysplastic Syndrome HSCs. Nat. Immunol. 2020, 21, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Saenz, D.T.; Fiskus, W.; Manshouri, T.; Rajapakshe, K.; Krieger, S.; Sun, B.; Mill, C.P.; DiNardo, C.; Pemmaraju, N.; Kadia, T.; et al. BET Protein Bromodomain Inhibitor-Based Combinations Are Highly Active against Post-Myeloproliferative Neoplasm Secondary AML Cells. Leukemia 2017, 31, 678–687. [Google Scholar] [CrossRef] [Green Version]
- Berthon, C.; Raffoux, E.; Thomas, X.; Vey, N.; Gomez-Roca, C.; Yee, K.; Taussig, D.C.; Rezai, K.; Roumier, C.; Herait, P.; et al. Bromodomain Inhibitor OTX015 in Patients with Acute Leukaemia: A Dose-Escalation, Phase 1 Study. Lancet Haematol. 2016, 3, e186–e195. [Google Scholar] [CrossRef]
- McCalmont, H.; Li, K.L.; Jones, L.; Toubia, J.; Bray, S.C.; Casolari, D.A.; Mayoh, C.; Samaraweera, S.E.; Lewis, I.D.; Prinjha, R.K.; et al. Efficacy of Combined CDK9/BET Inhibition in Preclinical Models of MLL-Rearranged Acute Leukemia. Blood Adv. 2020, 4, 296–300. [Google Scholar] [CrossRef]
- Mill, C.P.; Fiskus, W.; DiNardo, C.D.; Qian, Y.; Raina, K.; Rajapakshe, K.; Perera, D.; Coarfa, C.; Kadia, T.M.; Khoury, J.D.; et al. RUNX1-Targeted Therapy for AML Expressing Somatic or Germline Mutation in RUNX1. Blood 2019, 134, 59–73. [Google Scholar] [CrossRef]
- Shorstova, T.; Foulkes, W.D.; Witcher, M. Achieving Clinical Success with BET Inhibitors as Anti-Cancer Agents. Br. J. Cancer 2021, 124, 1478–1490. [Google Scholar] [CrossRef]
- Mill, C.P.; Fiskus, W.; DiNardo, C.D.; Birdwell, C.; Davis, J.A.; Kadia, T.M.; Takahashi, K.; Short, N.; Daver, N.; Ohanian, M.; et al. Effective Therapy for AML with RUNX1 Mutation by Cotreatment with Inhibitors of Protein Translation and BCL2. Blood 2022, 139, 907–921. [Google Scholar] [CrossRef]
- Du, X.; Chapman, N.M.; Chi, H. Emerging Roles of Cellular Metabolism in Regulating Dendritic Cell Subsets and Function. Front. Cell Dev. Biol. 2018, 6, 152. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Sanin, D.E.; Everts, B.; Chen, Q.; Qiu, J.; Buck, M.D.; Patterson, A.; Smith, A.M.; Chang, C.-H.; Liu, Z.; et al. Type 1 Interferons Induce Changes in Core Metabolism That Are Critical for Immune Function. Immunity 2016, 44, 1325–1336. [Google Scholar] [CrossRef] [Green Version]
- Qiu, C.C.; Lee, M.H.; Gallucci, S. Modulation of the Energy Metabolism Inhibits Plasmacytoid Dendritic Cell Activation and Delays Onset of Autoantibody Production in Murine Models of Systemic Lupus Erythematosus. J. Immunol. 2017, 198, 80.12. [Google Scholar]
- Ceroi, A.; Masson, D.; Roggy, A.; Roumier, C.; Chagué, C.; Gauthier, T.; Philippe, L.; Lamarthée, B.; Angelot-Delettre, F.; Bonnefoy, F.; et al. LXR Agonist Treatment of Blastic Plasmacytoid Dendritic Cell Neoplasm Restores Cholesterol Efflux and Triggers Apoptosis. Blood 2016, 128, 2694–2707. [Google Scholar] [CrossRef] [Green Version]

| Markers | Blastic pDC from BPDCN | pDC from MPDCP | Normal Mature pDC |
|---|---|---|---|
| lin (CD3, CD19, CD14, CD16, cMPO) | Negative | Negative | Negative |
| CD45RA | Positive | Positive | Positive |
| CD123 | High | High | High |
| HLA-DR | High | High | Positive |
| CD4 | Positive | Positive | Positive |
| CD303 | Intermediate | Positive | High |
| CD304 | Intermediate | Positive | High |
| CD11c | Negative | Negative | Negative |
| CD56 | Positive | Negative | Negative * |
| CD34 | Negative | Positive | Negative |
| CD5 | Positive/Negative | Positive/Negative | Negative * |
| CD7 | Positive/Negative | Positive/Negative | Negative |
| CD22 | Positive/Negative | Positive/Negative | Negative |
| TdT | Positive/Negative | Positive/Negative | Positive |
| cTCL-1 | High | Intermediate | Negative |
| nTCF4 | Positive | Positive | Positive |
| Therapeutic Class | Targets | Type of Therapy |
|---|---|---|
| Chemotherapy | Asparagine | L asparaginase |
| Adoptive therapies | pDC & blasts | ASCT |
| CD123 | mAb, CAR T-cells | |
| FLT3 | BiTE | |
| TAA | tgTCR, vaccination | |
| Immune therapies | PD-1 | Immune checkpoint molecule blockade |
| PD-L1 | ||
| CTLA-4 | ||
| GITR | ||
| ICOS | ||
| TIGIT | ||
| CD47 | ||
| Targeted therapies | BET | Inhibitors |
| MEK | ||
| BCL2 | ||
| JAK2 | ||
| IDO1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roussel, X.; Garnache Ottou, F.; Renosi, F. Plasmacytoid Dendritic Cells, a Novel Target in Myeloid Neoplasms. Cancers 2022, 14, 3545. https://doi.org/10.3390/cancers14143545
Roussel X, Garnache Ottou F, Renosi F. Plasmacytoid Dendritic Cells, a Novel Target in Myeloid Neoplasms. Cancers. 2022; 14(14):3545. https://doi.org/10.3390/cancers14143545
Chicago/Turabian StyleRoussel, Xavier, Francine Garnache Ottou, and Florian Renosi. 2022. "Plasmacytoid Dendritic Cells, a Novel Target in Myeloid Neoplasms" Cancers 14, no. 14: 3545. https://doi.org/10.3390/cancers14143545