Biomarkers for the Detection and Management of Hepatocellular Carcinoma in Patients Treated with Direct-Acting Antivirals
Abstract
:Simple Summary
Abstract
1. Introduction
2. Molecular Mechanism of HCC Development in HCV Patients
3. The Controversial Role of DAA Treatment in HCC Development
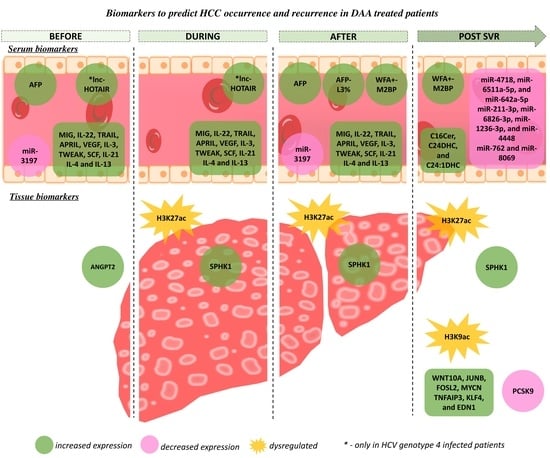
4. Predictive Biomarkers of HCC Development in DAA-Treated Patients
4.1. Tissue Biomarkers to Predict the Risk of HCC in DAA Treated Patients
4.1.1. Angiogenic Factors
4.1.2. Epigenetic Footprints
4.2. Serum Biomarkers to Predict the Risk of HCC in DAA-Treated Patients
4.2.1. Alpha-Fetoprotein
4.2.2. WFA+-M2BP
4.2.3. Circulating Immune Mediators
4.2.4. Non-Coding RNAs
4.2.5. Sphingolipids
5. Validation of Pre-Clinical Biomarkers
| Source | Biomarkers | Dysregulation in Relation to DAA Treatment | Risk Predictor of HCC | Cohort Size | Refs. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Before | During | After | Post SVR | Occurrence | Recurrence | ||||
| Tissue Biomarkers |
| ↑ | √ | √ | 242 | [75] | |||
| √ | -- | 48 | [76] | |||||
| ↑ | ↑ | ↑ | |||||||
| √ | -- | 17 | [74] | |||||
| ↑ | |||||||||
| ↓ | |||||||||
| Serum Biomarkers |
| ↑ | ↑ | √ | √ | 234 | [86] | ||
| ↑ | √ | √ | 220 | [86] | ||||
| ↑ | ↑ | √ | -- | 23 | [98] | |||
| ↑ | ↑ | ↑ | √ | -- | 49 | [69,93] | ||
| ↓ | ↓ | √ | -- | 60 | [101] | |||
| ↑ | √ | -- | 166 | [104] | ||||
| ↑ | ↑ | √ | -- | 567 | [87] | |||
| ↓ | -- | √ | 139 | [103] | ||||
| ↓ | √ | -- | 139 | [103] | ||||
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Petrick, J.L.; Florio, A.A.; Znaor, A.; Ruggieri, D.; Laversanne, M.; Alvarez, C.S.; Ferlay, J.; Valery, P.C.; Bray, F.; McGlynn, K.A. International Trends in Hepatocellular Carcinoma Incidence, 1978–2012. Int. J. Cancer 2020, 147, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.-L.; Schirmacher, P.; Viligrain, V. EASL Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [Green Version]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular Carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jie, W.; Ling, J.; Yuanshuai, H. HCV Core Antigen Plays an Important Role in the Fight against HCV as an Alternative to HCV-RNA Detection. J. Clin. Lab. Anal. 2021, 35, e23755. [Google Scholar] [CrossRef] [PubMed]
- Goossens, N.; Hoshida, Y. Hepatitis C Virus-Induced Hepatocellular Carcinoma. Clin. Mol. Hepatol. 2015, 21, 105–114. [Google Scholar] [CrossRef]
- Li, D.K.; Chung, R.T. Overview of Direct-Acting Antiviral Drugs and Drug Resistance of Hepatitis C Virus. Methods Mol. Biol. 2019, 1911, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Le Page, C.; Génin, P.; Baines, M.G.; Hiscott, J. Interferon Activation and Innate Immunity. Rev. Immunogenet. 2000, 2, 374–386. [Google Scholar]
- Leoni, M.C.; Ustianowski, A.; Farooq, H.; Arends, J.E. HIV, HCV and HBV: A Review of Parallels and Differences. Infect. Dis. Ther. 2018, 7, 407–419. [Google Scholar] [CrossRef] [Green Version]
- Chayama, K.; Imamura, M.; Hayes, C.N. Hepatitis C Virus Treatment Update—A New Era of All-Oral HCV Treatment. Adv. Dig. Med. 2016, 3, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.A.; Friedman, S.L. Reversal, Maintenance or Progression: What Happens to the Liver after a Virologic Cure of Hepatitis C? Antivir. Res. 2014, 107, 23–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aghemo, A.; Francesco, R.D. New Horizons in Hepatitis C Antiviral Therapy with Direct-Acting Antivirals. Hepatology 2013, 58, 428–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, S.B. The Possible Association Between DAA Treatment for HCV Infection and HCC Recurrence. Gastroenterol. Hepatol. 2016, 12, 776–779. [Google Scholar]
- Borgia, M.; Dal Bo, M.; Toffoli, G. Role of Virus-Related Chronic Inflammation and Mechanisms of Cancer Immune-Suppression in Pathogenesis and Progression of Hepatocellular Carcinoma. Cancers 2021, 13, 4387. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Lagaye, S. Hepatitis C Virus: Morphogenesis, Infection and Therapy. World J. Hepatol. 2018, 10, 186–212. [Google Scholar] [CrossRef] [PubMed]
- Hedskog, C.; Parhy, B.; Chang, S.; Zeuzem, S.; Moreno, C.; Shafran, S.D.; Borgia, S.M.; Asselah, T.; Alric, L.; Abergel, A.; et al. Identification of 19 Novel Hepatitis C Virus Subtypes—Further Expanding HCV Classification. Open Forum Infect. Dis. 2019, 6, ofz076. [Google Scholar] [CrossRef]
- Murphy, D.G.; Sablon, E.; Chamberland, J.; Fournier, E.; Dandavino, R.; Tremblay, C.L. Hepatitis C Virus Genotype 7, a New Genotype Originating from Central Africa. J. Clin. Microbiol. 2015, 53, 967–972. [Google Scholar] [CrossRef] [Green Version]
- Borgia, S.M.; Hedskog, C.; Parhy, B.; Hyland, R.H.; Stamm, L.M.; Brainard, D.M.; Subramanian, M.G.; McHutchison, J.G.; Mo, H.; Svarovskaia, E.; et al. Identification of a Novel Hepatitis C Virus Genotype From Punjab, India: Expanding Classification of Hepatitis C Virus Into 8 Genotypes. J. Infect. Dis. 2018, 218, 1722–1729. [Google Scholar] [CrossRef] [Green Version]
- Messina, J.P.; Humphreys, I.; Flaxman, A.; Brown, A.; Cooke, G.S.; Pybus, O.G.; Barnes, E. Global Distribution and Prevalence of Hepatitis C Virus Genotypes. Hepatology 2015, 61, 77–87. [Google Scholar] [CrossRef] [Green Version]
- Moradpour, D.; Penin, F. Hepatitis C Virus Proteins: From Structure to Function. Curr. Top. Microbiol. Immunol. 2013, 369, 113–142. [Google Scholar] [CrossRef]
- Saitta, C.; Tripodi, G.; Barbera, A.; Bertuccio, A.; Smedile, A.; Ciancio, A.; Raffa, G.; Sangiovanni, A.; Navarra, G.; Raimondo, G.; et al. Hepatitis B Virus (HBV) DNA Integration in Patients with Occult HBV Infection and Hepatocellular Carcinoma. Liver Int. 2015, 35, 2311–2317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukowati, C.H.C.; El-Khobar, K.E.; Ie, S.I.; Anfuso, B.; Muljono, D.H.; Tiribelli, C. Significance of Hepatitis Virus Infection in the Oncogenic Initiation of Hepatocellular Carcinoma. World J. Gastroenterol. 2016, 22, 1497–1512. [Google Scholar] [CrossRef] [PubMed]
- D’souza, S.; Lau, K.C.; Coffin, C.S.; Patel, T.R. Molecular Mechanisms of Viral Hepatitis Induced Hepatocellular Carcinoma. World J. Gastroenterol. 2020, 26, 5759–5783. [Google Scholar] [CrossRef]
- Mahmoudvand, S.; Shokri, S.; Taherkhani, R.; Farshadpour, F. Hepatitis C Virus Core Protein Modulates Several Signaling Pathways Involved in Hepatocellular Carcinoma. World J. Gastroenterol. 2019, 25, 42–58. [Google Scholar] [CrossRef]
- Banerjee, A.; Ray, R.B.; Ray, R. Oncogenic Potential of Hepatitis C Virus Proteins. Viruses 2010, 2, 2108–2133. [Google Scholar] [CrossRef]
- Jouvin-Marche, E.; Macek Jílková, Z.; Thelu, M.-A.; Marche, H.; Fugier, E.; Van Campenhout, N.; Hoang, X.S.; Marlu, A.; Sturm, N.; Callanan, M.; et al. Lymphocytes Degranulation in Liver in Hepatitis C Virus Carriers Is Associated With IFNL4 Polymorphisms and ALT Levels. J. Infect. Dis. 2014, 209, 1907–1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jilkova, Z.M.; Afzal, S.; Marche, H.; Decaens, T.; Sturm, N.; Jouvin-Marche, E.; Huard, B.; Marche, P.N. Progression of Fibrosis in Patients with Chronic Viral Hepatitis Is Associated with IL-17+ Neutrophils. Liver Int. 2016, 36, 1116–1124. [Google Scholar] [CrossRef]
- Haybaeck, J.; Zeller, N.; Wolf, M.J.; Weber, A.; Wagner, U.; Kurrer, M.O.; Bremer, J.; Iezzi, G.; Graf, R.; Clavien, P.-A.; et al. A Lymphotoxin-Driven Pathway to Hepatocellular Carcinoma. Cancer Cell 2009, 16, 295–308. [Google Scholar] [CrossRef] [Green Version]
- Ramzan, M.; Sturm, N.; Decaens, T.; Bioulac-Sage, P.; Bancel, B.; Merle, P.; Tran Van Nhieu, J.; Slama, R.; Letoublon, C.; Zarski, J.-P.; et al. Liver-Infiltrating CD8+ Lymphocytes as Prognostic Factor for Tumour Recurrence in Hepatitis C Virus-Related Hepatocellular Carcinoma. Liver Int. 2016, 36, 434–444. [Google Scholar] [CrossRef]
- Natural Killer T Cell—An Overview|ScienceDirect Topics. Available online: https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/natural-killer-t-cell (accessed on 6 May 2022).
- Vallianou, I.; Dafou, D.; Vassilaki, N.; Mavromara, P.; Hadzopoulou-Cladaras, M. Hepatitis C Virus Suppresses Hepatocyte Nuclear Factor 4 Alpha, a Key Regulator of Hepatocellular Carcinoma. Int. J. Biochem. Cell Biol. 2016, 78, 315–326. [Google Scholar] [CrossRef]
- Aydin, Y.; Kurt, R.; Song, K.; Lin, D.; Osman, H.; Youngquist, B.; Scott, J.W.; Shores, N.J.; Thevenot, P.; Cohen, A.; et al. Hepatic Stress Response in HCV Infection Promotes STAT3-Mediated Inhibition of HNF4A-MiR-122 Feedback Loop in Liver Fibrosis and Cancer Progression. Cancers 2019, 11, 1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dash, S.; Chava, S.; Aydin, Y.; Chandra, P.K.; Ferraris, P.; Chen, W.; Balart, L.A.; Wu, T.; Garry, R.F. Hepatitis C Virus Infection Induces Autophagy as a Prosurvival Mechanism to Alleviate Hepatic ER-Stress Response. Viruses 2016, 8, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, T.; Aburatani, H. Exploration of Liver Cancer Genomes. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, W.M.; Amr, K.S. Insights for Hepatitis C Virus Related Hepatocellular Carcinoma Genetic Biomarkers: Early Diagnosis and Therapeutic Intervention. World J. Hepatol. 2016, 8, 1251–1261. [Google Scholar] [CrossRef]
- Benegiamo, G.; Vinciguerra, M.; Mazzoccoli, G.; Piepoli, A.; Andriulli, A.; Pazienza, V. DNA Methyltransferases 1 and 3b Expression in Huh-7 Cells Expressing HCV Core Protein of Different Genotypes. Dig. Dis. Sci. 2012, 57, 1598–1603. [Google Scholar] [CrossRef]
- Duong, F.H.T.; Christen, V.; Lin, S.; Heim, M.H. Hepatitis C Virus-Induced up-Regulation of Protein Phosphatase 2A Inhibits Histone Modification and DNA Damage Repair. Hepatology 2010, 51, 741–751. [Google Scholar] [CrossRef]
- Abdallah, C.; Lejamtel, C.; Benzoubir, N.; Battaglia, S.; Sidahmed-Adrar, N.; Desterke, C.; Lemasson, M.; Rosenberg, A.R.; Samuel, D.; Bréchot, C.; et al. Hepatitis C Virus Core Protein Targets 4E-BP1 Expression and Phosphorylation and Potentiates Myc-Induced Liver Carcinogenesis in Transgenic Mice. Oncotarget 2017, 8, 56228–56242. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Pan, Q.; Fuhler, G.M.; Smits, R.; Peppelenbosch, M.P. Action and Function of Wnt/β-Catenin Signaling in the Progression from Chronic Hepatitis C to Hepatocellular Carcinoma. J. Gastroenterol. 2017, 52, 419–431. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, J.; Aoki, H.; Kajino, K.; Moriyama, M.; Arakawa, Y.; Hino, O. Hepatitis C Virus Core Protein Activates the MAPK/ERK Cascade Synergistically with Tumor Promoter TPA, but Not with Epidermal Growth Factor or Transforming Growth Factor α. Hepatology 2000, 32, 958–961. [Google Scholar] [CrossRef]
- Otsuka, M.; Kato, N.; Lan, K.-H.; Yoshida, H.; Kato, J.; Goto, T.; Shiratori, Y.; Omata, M. Hepatitis C Virus Core Protein Enhances P53 Function through Augmentation of DNA Binding Affinity and Transcriptional Ability. J. Biol. Chem. 2000, 275, 34122–34130. [Google Scholar] [CrossRef] [Green Version]
- Hassan, M.; Selimovic, D.; Ghozlan, H.; Abdel-kader, O. Hepatitis C Virus Core Protein Triggers Hepatic Angiogenesis by a Mechanism Including Multiple Pathways. Hepatology 2009, 49, 1469–1482. [Google Scholar] [CrossRef] [PubMed]
- Korenaga, M.; Wang, T.; Li, Y.; Showalter, L.A.; Chan, T.; Sun, J.; Weinman, S.A. Hepatitis C Virus Core Protein Inhibits Mitochondrial Electron Transport and Increases Reactive Oxygen Species (ROS) Production. J. Biol. Chem. 2005, 280, 37481–37488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florentin, J.; Aouar, B.; Dental, C.; Thumann, C.; Firaguay, G.; Gondois-Rey, F.; Soumelis, V.; Baumert, T.F.; Nunès, J.A.; Olive, D.; et al. HCV Glycoprotein E2 Is a Novel BDCA-2 Ligand and Acts as an Inhibitor of IFN Production by Plasmacytoid Dendritic Cells. Blood 2012, 120, 4544–4551. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhao, Y.; Gao, Y.; Hu, W.; Qu, Y.; Lou, N.; Zhu, Y.; Zhang, X.; Yang, H. Hepatitis C Virus NS3 Protein Enhances Hepatocellular Carcinoma Cell Invasion by Promoting PPM1A Ubiquitination and Degradation. J. Exp. Clin. Cancer Res. 2017, 36, 42. [Google Scholar] [CrossRef] [Green Version]
- Einav, S.; Sklan, E.H.; Moon, H.M.; Gehrig, E.; Liu, P.; Hao, Y.; Lowe, A.W.; Glenn, J.S. The Nucleotide Binding Motif of Hepatitis C Virus NS4B Can Mediate Cellular Transformation and Tumor Formation without Ha-Ras Co-Transfection. Hepatology 2008, 47, 827–835. [Google Scholar] [CrossRef]
- Cheng, D.; Zhao, L.; Zhang, L.; Jiang, Y.; Tian, Y.; Xiao, X.; Gong, G. P53 Controls Hepatitis C Virus Non-Structural Protein 5A-Mediated Downregulation of GADD45α Expression via the NF-ΚB and PI3K–Akt Pathways. J. Gen. Virol 2013, 94, 326–335. [Google Scholar] [CrossRef]
- Pascut, D.; Hoang, M.; Nguyen, N.N.Q.; Pratama, M.Y.; Tiribelli, C. HCV Proteins Modulate the Host Cell MiRNA Expression Contributing to Hepatitis C Pathogenesis and Hepatocellular Carcinoma Development. Cancers 2021, 13, 2485. [Google Scholar] [CrossRef]
- Singaravelu, R.; Chen, R.; Lyn, R.K.; Jones, D.M.; O’Hara, S.; Rouleau, Y.; Cheng, J.; Srinivasan, P.; Nasheri, N.; Russell, R.S.; et al. Hepatitis C Virus Induced Up-Regulation of MicroRNA-27: A Novel Mechanism for Hepatic Steatosis. Hepatology 2014, 59, 98–108. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Wang, H.; Shi, J.; Wu, K.; Liu, S.; Liu, Y.; Wu, J. HCV-Induced MiR-21 Contributes to Evasion of Host Immune System by Targeting MyD88 and IRAK1. PLoS Pathog. 2013, 9, e1003248. [Google Scholar] [CrossRef]
- Hung, C.-H. Insulin Resistance Is Associated with Hepatocellular Carcinoma in Chronic Hepatitis C Infection. WJG 2010, 16, 2265. [Google Scholar] [CrossRef]
- Shintani, Y.; Fujie, H.; Miyoshi, H.; Tsutsumi, T.; Tsukamoto, K.; Kimura, S.; Moriya, K.; Koike, K. Hepatitis C Virus Infection and Diabetes: Direct Involvement of the Virus in the Development of Insulin Resistance. Gastroenterology 2004, 126, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Romero-Gómez, M. Insulin Resistance and Hepatitis C. WJG 2006, 12, 7075. [Google Scholar] [CrossRef] [PubMed]
- Kralj, D.; Virović Jukić, L.; Stojsavljević, S.; Duvnjak, M.; Smolić, M.; Čurčić, I.B. Hepatitis C Virus, Insulin Resistance, and Steatosis. J. Clin. Transl. Hepatol. 2016, 4, 66–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, F.-K.F.; Torres, D.M.; Harrison, S.A. Hepatitis C and Lipid Metabolism, Hepatic Steatosis, and NAFLD: Still Important in the Era of Direct Acting Antiviral Therapy? J. Viral Hepat. 2014, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Suzuki, R.; Murakami, K.; Aizaki, H.; Ishii, K.; Murayama, A.; Date, T.; Matsuura, Y.; Miyamura, T.; Wakita, T.; et al. Interaction of Hepatitis C Virus Nonstructural Protein 5A with Core Protein Is Critical for the Production of Infectious Virus Particles. J. Virol. 2008, 82, 7964–7976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dharancy, S.; Malapel, M.; Perlemuter, G.; Roskams, T.; Cheng, Y.; Dubuquoy, L.; Podevin, P.; Conti, F.; Canva, V.; Philippe, D.; et al. Impaired Expression of the Peroxisome Proliferator–Activated Receptor Alpha during Hepatitis C Virus Infection. Gastroenterology 2005, 128, 334–342. [Google Scholar] [CrossRef] [Green Version]
- McPherson, S.; Jonsson, J.R.; Barrie, H.D.; O’Rourke, P.; Clouston, A.D.; Powell, E.E. Investigation of the Role of SREBP-1c in the Pathogenesis of HCV-Related Steatosis. J. Hepatol. 2008, 49, 1046–1054. [Google Scholar] [CrossRef]
- Paolucci, S.; Piralla, A.; Novazzi, F.; Fratini, A.; Maserati, R.; Gulminetti, R.; Novati, S.; Barbarini, G.; Sacchi, P.; Silvestri, A.D.; et al. Baseline Amino Acid Substitutions in the NS5A ISDR and PKR Binding Domain of Hepatitis C and Different Fibrosis Levels and Levels of Development of Hepatocellular Carcinoma in Patients Treated with DAAs. Viruses 2020, 12, 255. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Li, J.; Rupp, L.B.; Holmberg, S.D.; Moorman, A.C.; Spradling, P.R.; Teshale, E.H.; Zhou, Y.; Boscarino, J.A.; Schmidt, M.A.; et al. Hepatitis C Treatment Failure Is Associated with Increased Risk of Hepatocellular Carcinoma. J. Viral Hepat. 2016, 23, 718–729. [Google Scholar] [CrossRef]
- Fabregat, I.; Caballero-Díaz, D. Transforming Growth Factor-β-Induced Cell Plasticity in Liver Fibrosis and Hepatocarcinogenesis. Front. Oncol. 2018, 8, 357. [Google Scholar] [CrossRef] [Green Version]
- Chusri, P.; Kumthip, K.; Hong, J.; Zhu, C.; Duan, X.; Jilg, N.; Fusco, D.N.; Brisac, C.; Schaefer, E.A.; Cai, D.; et al. HCV Induces Transforming Growth Factor Β1 through Activation of Endoplasmic Reticulum Stress and the Unfolded Protein Response. Sci. Rep. 2016, 6, 22487. [Google Scholar] [CrossRef] [PubMed]
- Kotsiri, I.; Hadziyannis, E.; Georgiou, A.; Papageorgiou, M.-V.; Vlachogiannakos, I.; Papatheodoridis, G. Changes in Serum Transforming Growth Factor-Β1 Levels in Chronic Hepatitis C Patients under Antiviral Therapy. Ann. Gastroenterol. 2016, 29, 79–84. [Google Scholar] [PubMed]
- Kozbial, K.; Moser, S.; Schwarzer, R.; Laferl, H.; Al-Zoairy, R.; Stauber, R.; Stättermayer, A.F.; Beinhardt, S.; Graziadei, I.; Freissmuth, C.; et al. Unexpected High Incidence of Hepatocellular Carcinoma in Cirrhotic Patients with Sustained Virologic Response Following Interferon-Free Direct-Acting Antiviral Treatment. J. Hepatol. 2016, 65, 856–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.-C.; Tseng, K.-C.; Tzeng, I.-S.; Kao, J.-H. The Impact of Cytokine Change after Hepatitis C Virus Clearance by Direct Antiviral Agents on the Risk of Hepatocellular Carcinoma. J. Formos Med. Assoc. 2021, 120, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Reig, M.; Mariño, Z.; Perelló, C.; Iñarrairaegui, M.; Ribeiro, A.; Lens, S.; Díaz, A.; Vilana, R.; Darnell, A.; Varela, M.; et al. Unexpected High Rate of Early Tumor Recurrence in Patients with HCV-Related HCC Undergoing Interferon-Free Therapy. J. Hepatol. 2016, 65, 719–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, C.N.; Zhang, P.; Zhang, Y.; Chayama, K. Molecular Mechanisms of Hepatocarcinogenesis Following Sustained Virological Response in Patients with Chronic Hepatitis C Virus Infection. Viruses 2018, 10, 531. [Google Scholar] [CrossRef] [PubMed]
- Reig, M.; Boix, L.; Mariño, Z.; Torres, F.; Forns, X.; Bruix, J. Liver Cancer Emergence Associated with Antiviral Treatment: An Immune Surveillance Failure? Semin. Liver Dis. 2017, 37, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Macek Jílková, Z.; Seigneurin, A.; Coppard, C.; Ouaguia, L.; Aspord, C.; Marche, P.N.; Leroy, V.; Decaens, T. Circulating IL-13 Is Associated with de novo Development of HCC in HCV-Infected Patients Responding to Direct-Acting Antivirals. Cancers 2020, 12, 3820. [Google Scholar] [CrossRef]
- Ono, A.; Goossens, N.; Finn, R.S.; Schmidt, W.N.; Thung, S.N.; Im, G.Y.; Hoshida, Y. Persisting Risk of Hepatocellular Carcinoma after Hepatitis C Virus Cure Monitored by a Liver Transcriptome Signature. Hepatology 2017, 66, 1344–1346. [Google Scholar] [CrossRef] [Green Version]
- Guarino, M.; Sessa, A.; Cossiga, V.; Morando, F.; Caporaso, N.; Morisco, F. Direct-Acting Antivirals and Hepatocellular Carcinoma in Chronic Hepatitis C: A Few Lights and Many Shadows. World J. Gastroenterol. 2018, 24, 2582–2595. [Google Scholar] [CrossRef]
- Omland, L.H.; Krarup, H.; Jepsen, P.; Georgsen, J.; Harritshøj, L.H.; Riisom, K.; Jacobsen, S.E.H.; Schouenborg, P.; Christensen, P.B.; Sørensen, H.T.; et al. Mortality in Patients with Chronic and Cleared Hepatitis C Viral Infection: A Nationwide Cohort Study. J. Hepatol. 2010, 53, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, R.G.; Stasi, C. Recent Advancements in Diagnosis and Therapy of Liver Cirrhosis. Curr. Drug Targets 2016, 17, 1804–1817. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.; Kaspi, A.; Domovitz, T.; Davidovich, A.; Lavi-Itzkovitz, A.; Meirson, T.; Alison Holmes, J.; Dai, C.-Y.; Huang, C.-F.; Chung, R.T.; et al. Hepatitis C Virus Leaves an Epigenetic Signature Post Cure of Infection by Direct-Acting Antivirals. PLoS Genet. 2019, 15, e1008181. [Google Scholar] [CrossRef] [PubMed]
- Faillaci, F.; Marzi, L.; Critelli, R.; Milosa, F.; Schepis, F.; Turola, E.; Andreani, S.; Vandelli, G.; Bernabucci, V.; Lei, B.; et al. Liver Angiopoietin-2 Is a Key Predictor of de novo or Recurrent Hepatocellular Cancer After Hepatitis C Virus Direct-Acting Antivirals. Hepatology 2018, 68, 1010–1024. [Google Scholar] [CrossRef] [Green Version]
- Hamdane, N.; Jühling, F.; Crouchet, E.; Saghire, H.E.; Thumann, C.; Oudot, M.A.; Bandiera, S.; Saviano, A.; Ponsolles, C.; Suarez, A.A.R.; et al. HCV-Induced Epigenetic Changes Associated With Liver Cancer Risk Persist After Sustained Virologic Response. Gastroenterology 2019, 156, 2313–2329. [Google Scholar] [CrossRef] [Green Version]
- Villani, R.; Facciorusso, A.; Bellanti, F.; Tamborra, R.; Piscazzi, A.; Landriscina, M.; Vendemiale, G.; Serviddio, G. DAAs Rapidly Reduce Inflammation but Increase Serum VEGF Level: A Rationale for Tumor Risk during Anti-HCV Treatment. PLoS ONE 2016, 11, e0167934. [Google Scholar] [CrossRef] [Green Version]
- Goettsch, W.; Gryczka, C.; Korff, T.; Ernst, E.; Goettsch, C.; Seebach, J.; Schnittler, H.-J.; Augustin, H.G.; Morawietz, H. Flow-Dependent Regulation of Angiopoietin-2. J. Cell Physiol. 2008, 214, 491–503. [Google Scholar] [CrossRef]
- Kunz, P.; Hoffend, J.; Altmann, A.; Dimitrakopoulou-Strauss, A.; Koczan, D.; Eisenhut, M.; Bonaterra, G.A.; Dengler, T.J.; Mier, W.; Haberkorn, U.; et al. Angiopoietin-2 Overexpression in Morris Hepatoma Results in Increased Tumor Perfusion and Induction of Critical Angiogenesis-Promoting Genes. J. Nucl. Med. 2006, 47, 1515–1524. [Google Scholar]
- Jones, P.A.; Issa, J.-P.J.; Baylin, S. Targeting the Cancer Epigenome for Therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef]
- Lilley, C.E.; Chaurushiya, M.S.; Weitzman, M.D. Chromatin at the Intersection of Viral Infection and DNA Damage. Biochim. Et Biophys. Acta (BBA) Gene Regul. Mech. 2010, 1799, 319–327. [Google Scholar] [CrossRef] [Green Version]
- Rohrbach, T.; Maceyka, M.; Spiegel, S. Sphingosine Kinase and Sphingosine-1-Phosphate in Liver Pathobiology. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 543–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, H.; Xie, X.; Ji, L.; Ruan, X.; Zheng, Z. Sphingosine Kinase 1: A Novel Independent Prognosis Biomarker in Hepatocellular Carcinoma. Oncol. Lett. 2017, 13, 2316–2322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshida, Y.; Villanueva, A.; Sangiovanni, A.; Sole, M.; Hur, C.; Andersson, K.L.; Chung, R.T.; Gould, J.; Kojima, K.; Gupta, S.; et al. Prognostic Gene Expression Signature for Patients with Hepatitis C-Related Early-Stage Cirrhosis. Gastroenterology 2013, 144, 1024–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Zhang, K.-H. New Blood Biomarkers for the Diagnosis of AFP-Negative Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 1316. [Google Scholar] [CrossRef] [PubMed]
- Yoshimasu, Y.; Furuichi, Y.; Kasai, Y.; Takeuchi, H.; Sugimoto, K.; Nakamura, I.; Itoi, T. Predictive Factors for Hepatocellular Carcinoma Occurrence or Recurrence after Direct-Acting Antiviral Agents in Patients with Chronic Hepatitis C. J. Gastrointestin. Liver Dis. 2019, 28, 63–71. [Google Scholar] [CrossRef]
- Yasui, Y.; Kurosaki, M.; Komiyama, Y.; Takada, H.; Tamaki, N.; Watakabe, K.; Okada, M.; Wang, W.; Shimizu, T.; Kubota, Y.; et al. Wisteria Floribunda Agglutinin-Positive Mac-2 Binding Protein Predicts Early Occurrence of Hepatocellular Carcinoma after Sustained Virologic Response by Direct-Acting Antivirals for Hepatitis C Virus. Hepatol. Res. 2018, 48, 1131–1139. [Google Scholar] [CrossRef]
- Watanabe, T.; Tokumoto, Y.; Joko, K.; Michitaka, K.; Horiike, N.; Tanaka, Y.; Tada, F.; Kisaka, Y.; Nakanishi, S.; Yamauchi, K.; et al. Predictors of Hepatocellular Carcinoma Occurrence after Direct-Acting Antiviral Therapy in Patients with Hepatitis C Virus Infection. Hepatol. Res. 2019, 49, 136–146. [Google Scholar] [CrossRef]
- Lleo, A.; Aglitti, A.; Aghemo, A.; Maisonneuve, P.; Bruno, S.; Persico, M.; Rendina, M.; Ciancio, A.; Lampertico, P.; Brunetto, M.R.; et al. Predictors of Hepatocellular Carcinoma in HCV Cirrhotic Patients Treated with Direct Acting Antivirals. Dig. Liver Dis. 2019, 51, 310–317. [Google Scholar] [CrossRef]
- Bekki, Y.; Yoshizumi, T.; Shimoda, S.; Itoh, S.; Harimoto, N.; Ikegami, T.; Kuno, A.; Narimatsu, H.; Shirabe, K.; Maehara, Y. Hepatic Stellate Cells Secreting WFA+ -M2BP: Its Role in Biological Interactions with Kupffer Cells. J. Gastroenterol. Hepatol. 2017, 32, 1387–1393. [Google Scholar] [CrossRef]
- Strunz, B.; Hengst, J.; Deterding, K.; Manns, M.P.; Cornberg, M.; Ljunggren, H.-G.; Wedemeyer, H.; Björkström, N.K. Chronic Hepatitis C Virus Infection Irreversibly Impacts Human Natural Killer Cell Repertoire Diversity. Nat. Commun. 2018, 9, 2275. [Google Scholar] [CrossRef]
- Roche, B.; Coilly, A.; Duclos-Vallee, J.C.; Samuel, D. The Impact of Treatment of Hepatitis C with DAAs on the Occurrence of HCC. Liver Int. 2018, 38, 139–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debes, J.D.; van Tilborg, M.; Groothuismink, Z.M.A.; Hansen, B.E.; zur Wiesch, J.S.; von Felden, J.; de Knegt, R.J.; Boonstra, A. Levels of Cytokines in Serum Associate With Development of Hepatocellular Carcinoma in Patients With HCV Infection Treated With Direct-Acting Antivirals. Gastroenterology 2018, 154, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.-C.; Llovet, J.M. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015, 149, 1226–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panni, S.; Lovering, R.C.; Porras, P.; Orchard, S. Non-Coding RNA Regulatory Networks. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194417. [Google Scholar] [CrossRef] [PubMed]
- Ghai, V.; Lee, I.; Wang, K. Chapter 13—Circulating MiRNAs as Tumor Biomarkers. In Oncogenomics; Dammacco, F., Silvestris, F., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 191–206. ISBN 978-0-12-811785-9. [Google Scholar]
- Sukowati, C.H.C.; Cabral, L.K.D.; Tiribelli, C.; Pascut, D. Circulating Long and Circular Noncoding RNA as Non-Invasive Diagnostic Tools of Hepatocellular Carcinoma. Biomedicines 2021, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- El-Khazragy, N.; Elshimy, A.A.; Hassan, S.S.; Shaaban, M.H.; Bayoumi, A.H.; Magdoub, H.M.E.; Ghozy, S.; Gaballah, A.; Aboelhussein, M.M.; Gabal, H.H.A.; et al. Lnc-HOTAIR Predicts Hepatocellular Carcinoma in Chronic Hepatitis C Genotype 4 Following Direct-Acting Antivirals Therapy. Mol. Carcinog. 2020, 59, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, L.; Wu, L.-M.; Lai, M.-C.; Xie, H.-Y.; Zhang, F.; Zheng, S.-S. Overexpression of Long Non-Coding RNA HOTAIR Predicts Tumor Recurrence in Hepatocellular Carcinoma Patients Following Liver Transplantation. Ann. Surg. Oncol. 2011, 18, 1243–1250. [Google Scholar] [CrossRef]
- Yao, Y.; Li, J.; Wang, L. Large Intervening Non-Coding RNA HOTAIR Is an Indicator of Poor Prognosis and a Therapeutic Target in Human Cancers. Int. J. Mol. Sci. 2014, 15, 18985–18999. [Google Scholar] [CrossRef] [Green Version]
- Pascut, D.; Cavalletto, L.; Pratama, M.Y.; Bresolin, S.; Trentin, L.; Basso, G.; Bedogni, G.; Tiribelli, C.; Chemello, L. Serum MiRNA Are Promising Biomarkers for the Detection of Early Hepatocellular Carcinoma after Treatment with Direct-Acting Antivirals. Cancers 2019, 11, 1773. [Google Scholar] [CrossRef] [Green Version]
- Liese, J.; Peveling-Oberhag, J.; Doering, C.; Schnitzbauer, A.A.; Herrmann, E.; Zangos, S.; Hansmann, M.L.; Moench, C.; Welker, M.W.; Zeuzem, S.; et al. A Possible Role of MicroRNAs as Predictive Markers for the Recurrence of Hepatocellular Carcinoma after Liver Transplantation. Transpl. Int. 2016, 29, 369–380. [Google Scholar] [CrossRef] [Green Version]
- Itami-Matsumoto, S.; Hayakawa, M.; Uchida-Kobayashi, S.; Enomoto, M.; Tamori, A.; Mizuno, K.; Toyoda, H.; Tamura, T.; Akutsu, T.; Ochiya, T.; et al. Circulating Exosomal MiRNA Profiles Predict the Occurrence and Recurrence of Hepatocellular Carcinoma in Patients with Direct-Acting Antiviral-Induced Sustained Viral Response. Biomedicines 2019, 7, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mücke, V.T.; Thomas, D.; Mücke, M.M.; Waidmann, O.; Zeuzem, S.; Sarrazin, C.; Pfeilschifter, J.; Vermehren, J.; Finkelmeier, F.; Grammatikos, G. Serum Sphingolipids Predict de novo Hepatocellular Carcinoma in Hepatitis C Cirrhotic Patients with Sustained Virologic Response. Liver Int. 2019, 39, 2174–2183. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Parikh, N.D. Biomarker Development for Hepatocellular Carcinoma Early Detection: Current and Future Perspectives. Hepatic Oncol. 2017, 4, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Sherman, M. American Association for the Study of Liver Diseases Management of Hepatocellular Carcinoma: An Update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, H.; Yu, J. Biomarkers in Hepatocellular Carcinoma: Current Status and Future Perspectives. Biomedicines 2020, 8, 576. [Google Scholar] [CrossRef]

| DAAs | Components | Targets |
|---|---|---|
| Daklinza 1 | Daclatasvir | NS5A |
| Epclusa | Sofosbuvir | NS5B |
| Velpatasvir | NS5A | |
| Exviera | Dasabuvir | NS5B |
| Harvoni | Lepidasvir | NS5A |
| Sofosbuvir | NS5B | |
| Maviret | Glecaprevir | NS3/NS4A |
| Pibrentasvir | NS5A | |
| Olysio 1 | Simeprevir | NS3/NS4A |
| Sovaldi | Sofosbuvir | NS5B |
| Viekirax | Ombitasvir | NS5A |
| Paritaprevir | NS3/NS4A | |
| Ritonavir | NS3/NS4A | |
| Vosevi | Sofosbuvir | NS5B |
| Velpatasvir | NS5A | |
| Voxilaprevir | NS3/NS4A | |
| Zepatier | Elbasvir | NS5A |
| Grazoprevir | NS3/NS4A |
| Gene | Proteins | Functions |
|---|---|---|
| Structural proteins | ||
| Core (C) | p22 | Nucleocapsid |
| E1 | gp35 | Envelope glycoprotein |
| E2 | gp70 | Envelope glycoprotein |
| Non-structural proteins | ||
| NS1 | p7 | Short membrane peptide with possible ion channel function |
| NS2 | p23 | Cysteine protease |
| NS3 | p70 | Serine protease, RNA helicase with NTPase activity |
| NS4A | p8 | Cofactor for NS3 |
| NS4B | p27 | Integral protein inducing membranous web formation |
| NS5A | p56/p58 | Poly-phosphoprotein involved in HCV replication, modulation of cell signaling pathways, and mediation of IFN response |
| NS5B | p68 | RNA-dependent RNA polymerase |
| Pathway | Gene | HCV Protein | Reference |
|---|---|---|---|
| Angiogenesis | TGF-β2, VEGF | Core | [42] |
| Cell cycle regulation | -- | NS5A | [59] |
| Cell cycle regulation and DNA repair | p53 | Core | [41] |
| Cell cycle regulation and proliferation | MAPK/ERK pathway members | Core | [40] |
| EMT and invasion | PPM1A (ubiquitination) | NS3 | [45] |
| Epigenetic changes | DNMT1/DNMT3 | Core | [36] |
| IFN reduction | C-JUN, C-FOS, AP-1, miR-21, MyD88, IRAK1 | NS5ANS3/NS4A | [50] |
| Inflammation and cirrhosis progression | WNT/β-catenin pathway members | Core and NS5A | [39] |
| Immune escape | CLR, INFα/γ, Akt, ERK1/2 | E2 | [44] |
| Lipid accumulation | SREBP-1c | Core | [58] |
| Lipid metabolism and inflammation | PPARα | Core | [57] |
| Lipid metabolism and hepatic steatosis | miR-27a/27b, ANGPTL3, PPARα | NS4B | [49] |
| Oxidative stress | -- | Core | [43] |
| Proliferation | GADD45α | NS5A | [47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabral, L.K.D.; Grisetti, L.; Pratama, M.Y.; Tiribelli, C.; Pascut, D. Biomarkers for the Detection and Management of Hepatocellular Carcinoma in Patients Treated with Direct-Acting Antivirals. Cancers 2022, 14, 2700. https://doi.org/10.3390/cancers14112700
Cabral LKD, Grisetti L, Pratama MY, Tiribelli C, Pascut D. Biomarkers for the Detection and Management of Hepatocellular Carcinoma in Patients Treated with Direct-Acting Antivirals. Cancers. 2022; 14(11):2700. https://doi.org/10.3390/cancers14112700
Chicago/Turabian StyleCabral, Loraine Kay D., Luca Grisetti, Muhammad Yogi Pratama, Claudio Tiribelli, and Devis Pascut. 2022. "Biomarkers for the Detection and Management of Hepatocellular Carcinoma in Patients Treated with Direct-Acting Antivirals" Cancers 14, no. 11: 2700. https://doi.org/10.3390/cancers14112700