High Dual Expression of the Biomarkers CD44v6/α2β1 and CD44v6/PD-L1 Indicate Early Recurrence after Colorectal Hepatic Metastasectomy
Abstract
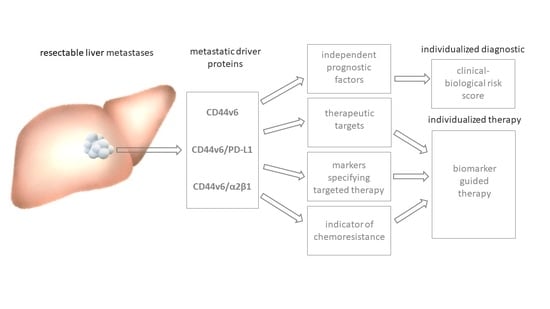
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Immunohistochemistry and Evaluation of Biomarker Expression
2.3. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Differential Biomarker Expression in Colorectal Liver and Lung Metastases
3.3. Prognostic Impact of Biomarker Expression in Colorectal Liver Metastases
3.4. CD44v6-Related Dual Biomarker Expression in Colorectal Liver Metastases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Farkas, L.; et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2021, 19, 329–359. [Google Scholar] [CrossRef] [PubMed]
- 2020 Exceptional Surveillance of Colorectal Cancer (NICE Guideline NG151); Copyright © NICE 2020; National Institute for Health and Care Excellence: London, UK, 2020.
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Nordlinger, B.; Sorbye, H.; Glimelius, B.; Poston, G.J.; Schlag, P.M.; Rougier, P.; Bechstein, W.O.; Primrose, J.N.; Walpole, E.T.; Finch-Jones, M.; et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): Long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013, 14, 1208–1215. [Google Scholar] [CrossRef]
- Kanemitsu, Y.; Shimizu, Y.; Mizusawa, J.; Inaba, Y.; Hamaguchi, T.; Shida, D.; Ohue, M.; Komori, K.; Shiomi, A.; Shiozawa, M.; et al. Hepatectomy Followed by mFOLFOX6 Versus Hepatectomy Alone for Liver-Only Metastatic Colorectal Cancer (JCOG0603): A Phase II or III Randomized Controlled Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 3789–3799. [Google Scholar] [CrossRef] [PubMed]
- Guerrera, F.; Falcoz, P.E.; Renaud, S.; Massard, G. Does perioperative chemotherapy improve survival in patients with resectable lung metastases of colorectal cancer? Interact. Cardiovasc. Thorac. Surg. 2017, 24, 789–791. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Qin, Y. Peri-operative chemotherapy for resectable colorectal lung metastasis: A systematic review and meta-analysis. J. Cancer. Res. Clin. Oncol. 2020, 146, 545–553. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.; Petrillo, A.; Smyth, E.C.; Shaida, N.; Khwaja, S.; Cheow, H.K.; Duckworth, A.; Heister, P.; Praseedom, R.; Jah, A.; et al. Colorectal liver metastases: Current management and future perspectives. World J. Clin. Oncol. 2020, 11, 761–808. [Google Scholar] [CrossRef]
- Lang, H.; Baumgart, J.; Roth, W.; Moehler, M.; Kloth, M. Cancer gene related characterization of patterns and point of recurrence after resection of colorectal liver metastases. Ann. Transl. Med. 2021, 9, 1372. [Google Scholar] [CrossRef]
- Filip, S.; Vymetalkova, V.; Petera, J.; Vodickova, L.; Kubecek, O.; John, S.; Cecka, F.; Krupova, M.; Manethova, M.; Cervena, K.; et al. Distant Metastasis in Colorectal Cancer Patients-Do We Have New Predicting Clinicopathological and Molecular Biomarkers? A Comprehensive Review. Int. J. Mol. Sci. 2020, 21, 5255. [Google Scholar] [CrossRef]
- Riihimaki, M.; Hemminki, A.; Sundquist, J.; Hemminki, K. Patterns of metastasis in colon and rectal cancer. Sci. Rep. 2016, 6, 29765. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Bado, I.; Wang, H.; Zhang, W.; Rosen, J.M.; Zhang, X.H. Metastasis Organotropism: Redefining the Congenial Soil. Dev. Cell 2019, 49, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Pretzsch, E.; Bösch, F.; Neumann, J.; Ganschow, P.; Bazhin, A.; Guba, M.; Werner, J.; Angele, M. Mechanisms of Metastasis in Colorectal Cancer and Metastatic Organotropism: Hematogenous versus Peritoneal Spread. J. Oncol. 2019, 2019, 7407190. [Google Scholar] [CrossRef] [PubMed]
- Chandra, R.; Karalis, J.D.; Liu, C.; Murimwa, G.Z.; Voth Park, J.; Heid, C.A.; Reznik, S.I.; Huang, E.; Minna, J.D.; Brekken, R.A. The Colorectal Cancer Tumor Microenvironment and Its Impact on Liver and Lung Metastasis. Cancers 2021, 13, 6206. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Imyanitov, E.; Kuligina, E. Molecular testing for colorectal cancer: Clinical applications. World J. Gastrointest. Oncol. 2021, 13, 1288–1301. [Google Scholar] [CrossRef]
- Bhullar, D.S.; Barriuso, J.; Mullamitha, S.; Saunders, M.P.; O’Dwyer, S.T.; Aziz, O. Biomarker concordance between primary colorectal cancer and its metastases. eBioMedicine 2019, 40, 363–374. [Google Scholar] [CrossRef] [Green Version]
- Testa, U.; Castelli, G.; Pelosi, E. Genetic Alterations of Metastatic Colorectal Cancer. Biomedicines 2020, 8, 414. [Google Scholar] [CrossRef]
- Diener, M.K.; Fichtner-Feigl, S. Biomarkers in colorectal liver metastases: Rising complexity and unknown clinical significance? Ann. Gastroenterol. Surg. 2021, 5, 477–483. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, X.; Wang, X.; Chen, Y.; Li, Z.; Yu, J.; Yang, W.; Mao, B.; Zhang, H.; Li, J.; et al. Genetic differences between lung metastases and liver metastases from left-sided microsatellite stable colorectal cancer: Next generation sequencing and clinical implications. Ann. Transl. Med. 2021, 9, 967. [Google Scholar] [CrossRef]
- Jiang, B.; Mu, Q.; Qiu, F.; Li, X.; Xu, W.; Yu, J.; Fu, W.; Cao, Y.; Wang, J. Machine learning of genomic features in organotropic metastases stratifies progression risk of primary tumors. Nat. Commun. 2021, 12, 6692. [Google Scholar] [CrossRef]
- Puccini, A.; Seeber, A.; Xiu, J.; Goldberg, R.M.; Soldato, D.; Grothey, A.; Shields, A.F.; Salem, M.E.; Battaglin, F.; Berger, M.D.; et al. Molecular differences between lymph nodes and distant metastases compared with primaries in colorectal cancer patients. NPJ Precis. Oncol. 2021, 5, 95. [Google Scholar] [CrossRef] [PubMed]
- Eide, P.W.; Moosavi, S.H.; Eilertsen, I.A.; Brunsell, T.H.; Langerud, J.; Berg, K.C.G.; Røsok, B.I.; Bjørnbeth, B.A.; Nesbakken, A.; Lothe, R.A.; et al. Metastatic heterogeneity of the consensus molecular subtypes of colorectal cancer. NPJ Genom. Med. 2021, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, M.; Bronsert, P.; Fichtner-Feigl, S.; Jud, A.; Schilling, O. Proteome biology of primary colorectal carcinoma and corresponding liver metastases. Neoplasia 2021, 23, 1240–1251. [Google Scholar] [CrossRef]
- Vigano, L.; Corleone, P.; Darwish, S.S.; Turri, N.; Famularo, S.; Viggiani, L.; Rimassa, L.; Del Fabbro, D.; Di Tommaso, L.; Torzilli, G. Hepatic and Extrahepatic Colorectal Metastases Have Discordant Responses to Systemic Therapy. Pathology Data from Patients Undergoing Simultaneous Resection of Multiple Tumor Sites. Cancers 2021, 13, 464. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.H.; Chen, Y.X.; Fang, J.Y. Comprehensive review of targeted therapy for colorectal cancer. Signal. Transduct. Target. 2020, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Siena, S.; Sartore-Bianchi, A.; Marsoni, S.; Hurwitz, H.I.; McCall, S.J.; Penault-Llorca, F.; Srock, S.; Bardelli, A.; Trusolino, L. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1108–1119. [Google Scholar] [CrossRef] [PubMed]
- Oliveres, H.; Pesántez, D.; Maurel, J. Lessons to Learn for Adequate Targeted Therapy Development in Metastatic Colorectal Cancer Patients. Int. J. Mol. Sci. 2021, 22, 5019. [Google Scholar] [CrossRef]
- Liang, R.; Lin, Y.; Yuan, C.L.; Liu, Z.H.; Li, Y.Q.; Luo, X.L.; Ye, J.Z.; Ye, H.H. High expression of estrogen-related receptor α is significantly associated with poor prognosis in patients with colorectal cancer. Oncol. Lett. 2018, 15, 5933–5939. [Google Scholar] [CrossRef] [Green Version]
- Ye, S.B.; Cheng, Y.K.; Zhang, L.; Wang, X.P.; Wang, L.; Lan, P. Prognostic value of estrogen receptor-α and progesterone receptor in curatively resected colorectal cancer: A retrospective analysis with independent validations. BMC Cancer 2019, 19, 933. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Dong, L.; Chang, P. CD44v6 engages in colorectal cancer progression. Cell Death Dis. 2019, 10, 30. [Google Scholar] [CrossRef]
- Li, C.; Zuo, D.; Liu, T.; Yin, L.; Li, C.; Wang, L. Prognostic and Clinicopathological Significance of MUC Family Members in Colorectal Cancer: A Systematic Review and Meta-Analysis. Gastroenterol Res. Pr. 2019, 2019, 2391670. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Wang, J.; Li, W.; Hao, X.; Hang, Q. Roles of Integrins in Gastrointestinal Cancer Metastasis. Front Mol. Biosci. 2021, 8, 708779. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Guo, S.; Li, Z.; Li, D.; Zhan, Q. High expression of HSP90 is associated with poor prognosis in patients with colorectal cancer. Peer J. 2019, 7, e7946. [Google Scholar] [CrossRef]
- Oliveira, A.F.; Bretes, L.; Furtado, I. Review of PD-1/PD-L1 Inhibitors in Metastatic dMMR/MSI-H Colorectal Cancer. Front Oncol. 2019, 9, 396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlueter, F.; Doetzer, K.; Pruefer, M.; Bazhin, A.V.; Werner, J.; Mayer, B. Integrating Routine Clinical Factors to Stratify Colorectal Cancer Patients with Liver and Lung Metastases for Immune Therapy. J. Cancer Sci. Clin. Ther. 2021, 5, 49–62. [Google Scholar] [CrossRef]
- Dotzer, K.; Schluter, F.; Schoenberg, M.B.; Bazhin, A.V.; von Koch, F.E.; Schnelzer, A.; Anthuber, S.; Grab, D.; Czogalla, B.; Burges, A.; et al. Immune Heterogeneity Between Primary Tumors and Corresponding Metastatic Lesions and Response to Platinum Therapy in Primary Ovarian Cancer. Cancers 2019, 11, 1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, B.; Lorenz, C.; Babic, R.; Jauch, K.W.; Schildberg, F.W.; Funke, I.; Johnson, J.P. Expression of leukocyte cell adhesion molecules on gastric carcinomas: Possible involvement of LFA-3 expression in the development of distant metastases. Int. J. Cancer 1995, 64, 415–423. [Google Scholar] [CrossRef]
- Rüschoff, J.; Dietel, M.; Baretton, G.; Arbogast, S.; Walch, A.; Monges, G.; Chenard, M.P.; Penault-Llorca, F.; Nagelmeier, I.; Schlake, W.; et al. HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch. Int. J. Pathol. 2010, 457, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Sieh, W.; Kobel, M.; Longacre, T.A.; Bowtell, D.D.; deFazio, A.; Goodman, M.T.; Hogdall, E.; Deen, S.; Wentzensen, N.; Moysich, K.B.; et al. Hormone-receptor expression and ovarian cancer survival: An Ovarian Tumor Tissue Analysis consortium study. Lancet Oncol. 2013, 14, 853–862. [Google Scholar] [CrossRef] [Green Version]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. Arch. Pathol. Lab. Med. 2018, 142, 1364–1382. [Google Scholar] [CrossRef] [Green Version]
- Nagai, S.; Takenaka, K.; Sonobe, M.; Ogawa, E.; Wada, H.; Tanaka, F. A novel classification of MUC1 expression is correlated with tumor differentiation and postoperative prognosis in non-small cell lung cancer. J. Thorac. Oncol. 2006, 1, 46–51. [Google Scholar]
- Zeng, Y.; Zhang, Q.; Zhang, Y.; Lu, M.; Liu, Y.; Zheng, T.; Feng, S.; Hao, M.; Shi, H. MUC1 Predicts Colorectal Cancer Metastasis: A Systematic Review and Meta-Analysis of Case Controlled Studies. PLoS ONE 2015, 10, e0138049. [Google Scholar] [CrossRef] [PubMed]
- Korehisa, S.; Oki, E.; Iimori, M.; Nakaji, Y.; Shimokawa, M.; Saeki, H.; Okano, S.; Oda, Y.; Maehara, Y. Clinical significance of programmed cell death-ligand 1 expression and the immune microenvironment at the invasive front of colorectal cancers with high microsatellite instability. Int. J. Cancer 2018, 142, 822–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baran, B.; Mert Ozupek, N.; Yerli Tetik, N.; Acar, E.; Bekcioglu, O.; Baskin, Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterol. Res. 2018, 11, 264–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zihui Yong, Z.; Ching, G.T.H.; Ching, M.T.C. Metastatic Profile of Colorectal Cancer: Interplay Between Primary Tumor Location and KRAS Status. J. Surg. Res. 2020, 246, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Li, X.; Pan, Z.; Zhao, L.; Ding, Y. Comparison of clinicopathological features and KRAS gene mutation of left-sided and right-sided colon cancers. Int. J. Clin. Exp. Pathol. 2017, 10, 11353–11359. [Google Scholar] [PubMed]
- Takahashi, Y.; Sugai, T.; Habano, W.; Ishida, K.; Eizuka, M.; Otsuka, K.; Sasaki, A.; Takayuki, M.; Morikawa, T.; Unno, M.; et al. Molecular differences in the microsatellite stable phenotype between left-sided and right-sided colorectal cancer. Int. J. Cancer 2016, 139, 2493–2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brulé, S.Y.; Jonker, D.J.; Karapetis, C.S.; O’Callaghan, C.J.; Moore, M.J.; Wong, R.; Tebbutt, N.C.; Underhill, C.; Yip, D.; Zalcberg, J.R.; et al. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur. J. Cancer 2015, 51, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Welch, D.R.; Hurst, D.R. Defining the Hallmarks of Metastasis. Cancer Res. 2019, 79, 3011–3027. [Google Scholar] [CrossRef]
- Dmello, R.S.; To, S.Q.; Chand, A.L. Therapeutic Targeting of the Tumour Microenvironment in Metastatic Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 2067. [Google Scholar] [CrossRef]
- Altorki, N.K.; Markowitz, G.J.; Gao, D.; Port, J.L.; Saxena, A.; Stiles, B.; McGraw, T.; Mittal, V. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat. Rev. Cancer 2019, 19, 9–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, J.; Tian, H.; Zhang, F.; Zhang, Z.; Li, J.; Liu, X.; Li, X.; Liu, J.; Li, X.; Jin, D.; et al. Extracellular vesicles of carcinoma-associated fibroblasts creates a pre-metastatic niche in the lung through activating fibroblasts. Mol. Cancer 2019, 18, 175. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Zubeldia, I.; Dotor, J.; Redrado, M.; Bleau, A.M.; Manrique, I.; de Aberasturi, A.L.; Villalba, M.; Calvo, A. Co-migration of colon cancer cells and CAFs induced by TGFβ₁ enhances liver metastasis. Cell Tissue Res. 2015, 359, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Pan, C.; Hu, H.; Zheng, S.; Ding, L. Osteopontin-enhanced hepatic metastasis of colorectal cancer cells. PLoS ONE 2012, 7, e47901. [Google Scholar] [CrossRef] [Green Version]
- Naba, A.; Clauser, K.R.; Whittaker, C.A.; Carr, S.A.; Tanabe, K.K.; Hynes, R.O. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer 2014, 14, 518. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef]
- Passot, G.; Kim, B.J.; Glehen, O.; Mehran, R.J.; Kopetz, S.E.; Goere, D.; Overman, M.J.; Pocard, M.; Marchal, F.; Conrad, C.; et al. Impact of RAS Mutations in Metastatic Colorectal Cancer After Potentially Curative Resection: Does Site of Metastases Matter? Ann. Surg. Oncol. 2018, 25, 179–187. [Google Scholar] [CrossRef]
- García-Mulero, S.; Alonso, M.H.; Pardo, J.; Santos, C.; Sanjuan, X.; Salazar, R.; Moreno, V.; Piulats, J.M.; Sanz-Pamplona, R. Lung metastases share common immune features regardless of primary tumor origin. J. Immunother. Cancer 2020, 8, 491. [Google Scholar] [CrossRef]
- Wang, J.L.; Su, W.Y.; Lin, Y.W.; Xiong, H.; Chen, Y.X.; Xu, J.; Fang, J.Y. CD44v6 overexpression related to metastasis and poor prognosis of colorectal cancer: A meta-analysis. Oncotarget 2017, 8, 12866–12876. [Google Scholar] [CrossRef] [Green Version]
- Viganò, L.; Capussotti, L.; Lapointe, R.; Barroso, E.; Hubert, C.; Giuliante, F.; Ijzermans, J.N.; Mirza, D.F.; Elias, D.; Adam, R. Early recurrence after liver resection for colorectal metastases: Risk factors, prognosis, and treatment. A LiverMetSurvey-based study of 6025 patients. Ann. Surg. Oncol. 2014, 21, 1276–1286. [Google Scholar] [CrossRef]
- Dai, S.; Ye, Y.; Kong, X.; Li, J.; Ding, K. A predictive model for early recurrence of colorectal-cancer liver metastases based on clinical parameters. Gastroenterol. Rep. 2021, 9, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Guo, H.; Downey, L.; Marroquin, C.; Wei, J.; Kuo, P.C. Osteopontin-dependent CD44v6 expression and cell adhesion in HepG2 cells. Carcinogenesis 2003, 24, 1871–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, T.; Gross, W.; Zöller, M. CD44v6 coordinates tumor matrix-triggered motility and apoptosis resistance. J. Biol. Chem. 2011, 286, 15862–15874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naci, D.; Vuori, K.; Aoudjit, F. Alpha2beta1 integrin in cancer development and chemoresistance. Semin Cancer Biol. 2015, 35, 145–153. [Google Scholar] [CrossRef]
- Lee, Y.; Shin, J.H.; Longmire, M.; Wang, H.; Kohrt, H.E.; Chang, H.Y.; Sunwoo, J.B. CD44+ Cells in Head and Neck Squamous Cell Carcinoma Suppress T-Cell-Mediated Immunity by Selective Constitutive and Inducible Expression of PD-L1. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 3571–3581. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.C.; Liao, T.T.; Yang, M.H. Immune Adaptation of Colorectal Cancer Stem Cells and Their Interaction With the Tumor Microenvironment. Front. Oncol. 2020, 10, 588542. [Google Scholar] [CrossRef]
- Chen, K.; Li, Z.; Jiang, P.; Zhang, X.; Zhang, Y.; Jiang, Y.; He, Y.; Li, X. Co-expression of CD133, CD44v6 and human tissue factor is associated with metastasis and poor prognosis in pancreatic carcinoma. Oncol. Rep. 2014, 32, 755–763. [Google Scholar] [CrossRef] [Green Version]
- Ueda, A.; Takasawa, A.; Akimoto, T.; Takasawa, K.; Aoyama, T.; Ino, Y.; Nojima, M.; Ono, Y.; Murata, M.; Osanai, M.; et al. Prognostic significance of the co-expression of EGFR and HER2 in adenocarcinoma of the uterine cervix. PLoS ONE 2017, 12, e0184123. [Google Scholar] [CrossRef]
- Margonis, G.A.; Sasaki, K.; Gholami, S.; Kim, Y.; Andreatos, N.; Rezaee, N.; Deshwar, A.; Buettner, S.; Allen, P.J.; Kingham, T.P.; et al. Genetic And Morphological Evaluation (GAME) score for patients with colorectal liver metastases. Br. J. Surg. 2018, 105, 1210–1220. [Google Scholar] [CrossRef]
- Chen, Y.; Chang, W.; Ren, L.; Chen, J.; Tang, W.; Liu, T.; Jian, M.; Liu, Y.; Wei, Y.; Xu, J. Comprehensive Evaluation of Relapse Risk (CERR) Score for Colorectal Liver Metastases: Development and Validation. Oncologist 2020, 25, e1031–e1041. [Google Scholar] [CrossRef] [Green Version]
- Lourenço, B.N.; Pereira, R.F.; Barrias, C.C.; Fischbach, C.; Oliveira, C.; Granja, P.L. Engineering Modular Half-Antibody Conjugated Nanoparticles for Targeting CD44v6-Expressing Cancer Cells. Nanomaterials 2021, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Porcellini, S.; Asperti, C.; Corna, S.; Cicoria, E.; Valtolina, V.; Stornaiuolo, A.; Valentinis, B.; Bordignon, C.; Traversari, C. CAR T Cells Redirected to CD44v6 Control Tumor Growth in Lung and Ovary Adenocarcinoma Bearing Mice. Front. Immunol. 2020, 11, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shek, D.; Akhuba, L.; Carlino, M.S.; Nagrial, A.; Moujaber, T.; Read, S.A.; Gao, B.; Ahlenstiel, G. Immune-Checkpoint Inhibitors for Metastatic Colorectal Cancer: A Systematic Review of Clinical Outcomes. Cancers 2021, 13, 4345. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, J.; Grothey, A.; Van Cutsem, E.; Yaeger, R.; Wasan, H.; Yoshino, T.; Desai, J.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib Plus Cetuximab as a New Standard of Care for Previously Treated BRAF V600E-Mutant Metastatic Colorectal Cancer: Updated Survival Results and Subgroup Analyses from the BEACON Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 273–284. [Google Scholar] [CrossRef]
- Strickler, J.H.; Rushing, C.N.; Uronis, H.E.; Morse, M.A.; Niedzwiecki, D.; Blobe, G.C.; Moyer, A.N.; Bolch, E.; Webb, R.; Haley, S.; et al. Cabozantinib and Panitumumab for RAS Wild-Type Metastatic Colorectal Cancer. Oncologist 2021, 26, 465-e917. [Google Scholar] [CrossRef]
- Ooki, A.; Shinozaki, E.; Yamaguchi, K. Immunotherapy in Colorectal Cancer: Current and Future Strategies. J. Anus Rectum Colon 2021, 5, 11–24. [Google Scholar] [CrossRef]
- Pereira, C.; Ferreira, D.; Mendes, N.; Granja, P.L.; Almeida, G.M.; Oliveira, C. Expression of CD44v6-Containing Isoforms Influences Cisplatin Response in Gastric Cancer Cells. Cancers 2020, 12, 858. [Google Scholar] [CrossRef] [Green Version]
- Kamphues, C.; Beyer, K.; Margonis, G.A. Prognostic and Therapeutic Implications of Tumor Biology in Colorectal Liver Metastases. Cancers 2021, 14, 88. [Google Scholar] [CrossRef]



| Biomarker | Antibody/Clone | Species | Isotype | Working Concentration (µg/mL) | Kit | Source |
|---|---|---|---|---|---|---|
| HGF-R | Sp44 | rabbit | IgG1 | 2.12 | - | Spring Bioscience/Biomol, Pleasanton, CA, USA |
| IGF1-R | 24–31 | mouse | IgG1 | 4.0 | + | Invitrogen, Carlsbad, CA, USA |
| EGR-R | H11 | mouse | IgG1 | 2.94 | - | Dako, Santa Clara, CA, USA |
| Her2/neu | 4B5 | rabbit | IgG1 | 1.5 | - | Ventana, Roche, Basel, Switzerland |
| Erα | ID5 | mouse | IgG1 | 2.5 | + | Dako |
| PR | PgR 636 | mouse | IgG1 | 2.5 | + | Dako |
| Muc1 | Ma55.2 | mouse | IgG1 | 0.5 | - | Monosan, Uden, The Netherlands |
| CD44v6 | VFF-18 | mouse | IgG1 | 1.0 | - | eBioscience Affymetrix |
| α2β1 | BHA2.1 | mouse | IgG1 | 2.5 | - | Millipore, Burlington, MA, USA |
| Hsp90 | AC88 | mouse | IgG1 | 10.0 | + | Abcam, Cambridge, UK |
| PD-L1 | MIH1 | mouse | IgG1 | 10.0 | + | Affymetrix |
| Positive controls | ||||||
| Epcam | Ber-EP4 | mouse | IgG1 | 5.0 | - | Dako |
| Pan Cytokeratin | KL-1 | mouse | IgG1 | 0.32 | - | Zytomed Systems |
| isotype controls | ||||||
| MOPC-21 | MOPC-21 | mouse | IgG1 | 5.0 | - | Sigma-Aldrich, St. Louis, MO, USA |
| MOPC-21 | MOPC-21 | mouse | IgG1 | 4.0 | + | Sigma-Aldrich |
| MOPC-21 | MOPC-21 | mouse | IgG1 | 10.0 | + | Sigma-Aldrich |
| Rabbit mAb | DA1E | rabbit | IgG1 | 2.12 | - | Cell Signaling, Danvers, MA, USA |
| Parameters | Liver Metastases | Lung Metastases | ||
|---|---|---|---|---|
| n | % | n | % | |
| patient related | ||||
| sex | ||||
| male | 34 | 64.15 | 12 | 80.00 |
| female | 19 | 35.85 | 3 | 20.00 |
| age (years) | ||||
| median | 64 | 62 | ||
| mean | 64 | 59 | ||
| range | 30–89 | 37–74 | ||
| metastasis related | ||||
| grading | ||||
| G1/G2 | 39 | 81.25 | 11 | 73.33 |
| G3 | 9 | 18.75 | 4 | 26.67 |
| missing | 5 | 0 | ||
| number of metastases * | ||||
| 1 | 19 | 35.85 | 7 | 46.67 |
| >1 | 34 | 64.15 | 8 | 53.33 |
| diameter of the largest metastases (cm) | ||||
| median | 3.5 | 1.8 | ||
| mean | 4.29 | 2.25 | ||
| range | 1.3–21.7 | 0.9–3.3 | ||
| type of metastasis | ||||
| synchronous | 35 | 66.04 | 0 | 0.00 |
| metachronous | 18 | 33.6 | 15 | 100.00 |
| R-status | ||||
| R0 | 39 | 73.58 | 12 | 80.00 |
| R1 | 14 | 26.42 | 3 | 20.00 |
| distinction of metastasis | ||||
| unilobular | 23 | 43.4 | 5 | 33.33 |
| multilobular | 30 | 56.6 | 10 | 66.67 |
| anatomical site | ||||
| left sided | 7 | 13.21 | 7 | 46.67 |
| right sided | 15 | 28.30 | 8 | 53.33 |
| both sided | 31 | 58.49 | ||
| neoadjuvant chemotherapy # | ||||
| yes | 23 | 43.40 | 8 | 53.33 |
| no | 30 | 56.60 | 7 | 46.67 |
| therapy options | ||||
| oxaliplatin-based | 11 | 47.83 | 1 | 12.5 |
| irinotecan-based | 7 | 30.43 | 5 | 62.5 |
| others | 5 | 21.74 | 2 | 25.0 |
| Biomarker | Number of Positive Lesions | Number of Positive Cancer Cells (%) | Number of Positive Lesions above Cut-Offs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver | Lung | Median | Mean | Liver | Lung | |||||||||
| n = 53 | % | n = 15 | % | Liver | Lung | p-Value | Liver | Lung | Cut Off * | n = 53 | % | n = 15 | % | |
| HGF-R | 52 | 98.1 | 15 | 100 | 95 | 95 | 0.166 | 87.7 | 95.3 | n.t. | ||||
| IGF-1R | 50 | 94.3 | 15 | 100 | 90 | 100 | 0.013 | 75.8 | 92.3 | >80 | 29 | 54.7 | 12 | 80 |
| EGF-R | 45 | 84.9 | 15 | 100 | 40 | 70 | 0.004 | 41.5 | 68.0 | >50 | 25 | 47.2 | 12 | 80 |
| Her2/neu | 19 | 35.8 | 8 | 53.3 | 0 | 1 | 0.575 | 5.7 | 1.7 | >50 | 1 | 1.9 | 0 | 0 |
| ERα | 1 | 1.9 | 0 | 0 | 0 | 0 | n.t. | 0.6 | 0 | ≥1 | n.t. | |||
| PR | 0 | 0 | 0 | 0 | 0 | 0 | n.t. | 0 | 0 | ≥1 | n.t. | |||
| Muc1 | 26 | 49.1 | 9 | 60 | 0 | 1 | 0.614 | 6.8 | 5.9 | +/− | 26 | 49.1 | 9 | 60 |
| CD44v6 | 45 | 84.9 | 15 | 100 | 30 | 60 | 0.019 | 34.9 | 55.7 | >30 | 23 | 43.4 | 10 | 66.7 |
| α2β1 | 46 | 86.8 | 15 | 100 | 70 | 90 | 0.001 | 58.5 | 88.3 | >80 | 20 | 37.7 | 11 | 73.3 |
| Hsp90 | 51 | 96.2 | 15 | 100 | 75 | 80 | 0.475 | 68.7 | 73.9 | >70 | 26 | 49.1 | 9 | 60 |
| PD-L1 | 24 | 45.3 | 13 | 86.7 | 0 | 1 | 0.005 | 6.1 | 3.25 | >1 | 24 | 45.3 | 11 | 73.3 |
| Variable | Groups | Cox Regression | ||
|---|---|---|---|---|
| HR | p-Value | 95% CI | ||
| age (median in years) | >64/≤64 | 1.424 | 0.357 | 0.671–3.021 |
| number of metastases * | >1/≤1 | 1.221 | 0.572 | 0.610–2.454 |
| type of metastases | synchronous/metachronous | 4.206 | 0.004 | 1.572–11.254 |
| CD44v6 expression | >30%/≤30% | 2.369 | 0.016 | 1.175–4.777 |
| Combination | Number of Patients (n) | Log Rank p-Value | Median PFS (month) |
|---|---|---|---|
| CD44v6 high * | 22 | 0.01 | 7 |
| CD44v6 low | 30 | 15.5 | |
| CD44v6 high/IGF1-R high | 15 | 0.142 | 7 |
| CD44v6 high/IGF1-R low or CD44v6 low/IGF1-R high | 20 | 9 | |
| CD44v6 low/IGF1-R low | 17 | 17 | |
| CD44v6 high/EGF-R high | 11 | 0.217 | 6 |
| CD44v6 high/EGF-R low or CD44v6 low/EGF-R high | 24 | 11.5 | |
| CD44v6 low/EGF-R low | 17 | 9 | |
| CD44v6 high/Muc1 high | 11 | 0.574 | 8 |
| CD44v6 high/Muc1 low or CD44v6 low/Muc1 high | 23 | 11 | |
| CD44v6 high/Muc1 low | 18 | 7.5 | |
| CD44v6 high/α2β1 high | 11 | 0.002 | 3 |
| CD44v6 high/α2β1 low or CD44v6 low/α2β1 high | 18 | 9 | |
| CD44v6 low/α2β1 low | 23 | 24 | |
| CD44v6 high/Hsp90 high | 14 | 0.022 | 7 |
| CD44v6 high/Hsp90 low or CD44v6 low/Hsp90 high | 21 | 9 | |
| CD44v6 low/Hsp90 low | 17 | 17 | |
| CD44v6 high/PD-L1 high | 12 | 0.023 | 7 |
| CD44v6 high/PD-L1 low or CD44v6 low/PD-L1 high | 21 | 14 | |
| CD44v6 low/PD-L1 low | 19 | 11 |
| Variable | Groups | Cox Regression (PFS) | ||
|---|---|---|---|---|
| HR | p-Value | 95% CI | ||
| age (median in years) | >64/≤64 | 1.561 | 0.256 | 0.724–3.366 |
| number of metastases * | >1/≤1 | 1.398 | 0.358 | 0.684–2.855 |
| type of metastases | synchronous/metachronous | 3.813 | 0.008 | 1.407–10.332 |
| CD44v6/α2β1 expression | high/high vs. low/low | 4.135 | 0.002 | 1.648–10.375 |
| high/low and low/high vs. low/low | 1.784 | 0.145 | 0.819–3.886 | |
| age (median in years) | >64/≤64 | 1.129 | 0.773 | 0.496–2.568 |
| number of metastases | >1/≤1 | 1.321 | 0.460 | 0.632–2.762 |
| type of metastases | synchronous/metachronous | 3.345 | 0.013 | 1.289–8.680 |
| CD44v6/Hsp90 expression | high/high vs. low/low | 2.039 | 0.085 | 0.906–4.586 |
| high/low and low/high vs. low/low | 1.412 | 0.443 | 0.585–3.404 | |
| age (median in years) | >64/≤64 | 1.290 | 0.493 | 0.623–2.675 |
| number of metastases | >1/≤1 | 1.341 | 0.418 | 0.659–2.728 |
| type of metastases | synchronous/metachronous | 4.154 | 0.004 | 1.584–10.893 |
| CD44v6/PD-L1 expression | high/high vs. low/low | 2.882 | 0.017 | 1.213–6.848 |
| high/low and low/high vs. low/low | 0.872 | 0.723 | 0.409–1.860 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wrana, F.; Dötzer, K.; Prüfer, M.; Werner, J.; Mayer, B. High Dual Expression of the Biomarkers CD44v6/α2β1 and CD44v6/PD-L1 Indicate Early Recurrence after Colorectal Hepatic Metastasectomy. Cancers 2022, 14, 1939. https://doi.org/10.3390/cancers14081939
Wrana F, Dötzer K, Prüfer M, Werner J, Mayer B. High Dual Expression of the Biomarkers CD44v6/α2β1 and CD44v6/PD-L1 Indicate Early Recurrence after Colorectal Hepatic Metastasectomy. Cancers. 2022; 14(8):1939. https://doi.org/10.3390/cancers14081939
Chicago/Turabian StyleWrana, Friederike, Katharina Dötzer, Martin Prüfer, Jens Werner, and Barbara Mayer. 2022. "High Dual Expression of the Biomarkers CD44v6/α2β1 and CD44v6/PD-L1 Indicate Early Recurrence after Colorectal Hepatic Metastasectomy" Cancers 14, no. 8: 1939. https://doi.org/10.3390/cancers14081939