Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosomes Rescue Testicular Aging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
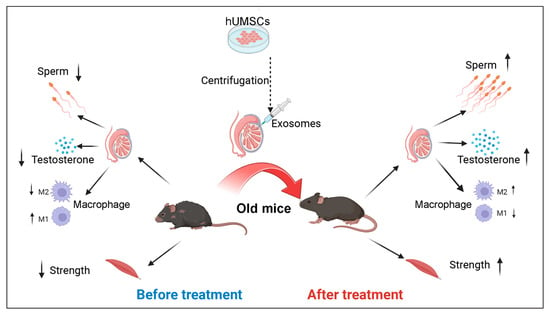
2.2. Study Design and Experimental Procedure
2.3. Culturing of hUMSCs
2.4. Preparation of Exosomes
2.5. Identification and Characterization of Exosomes
2.5.1. Transmission Electron Microscopy
2.5.2. Nanoparticle Tracking Analysis
2.5.3. Western Blotting
2.6. Exosomal Labeling In Vitro
2.7. SA-β-gal Assay
2.8. Testosterone Concentration Measurement
2.9. Hematoxylin and Eosin (H&E) Staining
2.10. Grip Strength Measurement
2.11. Immunofluorescence Staining
2.12. Computer-Aided Sperm Analysis (CASA)
2.13. Analysis of Macrophage Phenotype Using Flow Cytometry
2.14. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.15. Reactive Oxygen Species (ROS) Analysis
2.16. Bioinformatic Analysis
2.17. Statistical Analysis
3. Results
3.1. Isolation and Characterization of hUMSC-Exos
3.2. hUMSC-Exos Treatment Ameliorates Cellular Senescence in Aged Testis
3.3. hUMSC-Exos Treatment Improves Spermatogenesis in Aged Mice
3.4. hUMSC-Exos Treatment Promotes Testosterone Secretion in Aged Mice
3.5. hUMSC-Exos Treatment Modulates Macrophage Polarization
3.6. Bioinformatic Analysis of hUMSC-Exos miRNA Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaul, E.; Barron, J. Age-Related Diseases and Clinical and Public Health Implications for the 85 Years Old and Over Population. Front. Public Health 2017, 5, 335. [Google Scholar] [CrossRef] [PubMed]
- Frungieri, M.B.; Calandra, R.S.; Bartke, A.; Matzkin, M.E. Male and Female Gonadal Ageing: Its Impact on Health Span and Life Span. Mech. Ageing Dev. 2021, 197, 111519. [Google Scholar] [CrossRef] [PubMed]
- Broekmans, F.J.; Soules, M.R.; Fauser, B.C. Ovarian Aging: Mechanisms and Clinical Consequences. Endocr. Rev. 2009, 30, 465–493. [Google Scholar] [CrossRef] [PubMed]
- Huhtaniemi, I.; Forti, G. Male Late-Onset Hypogonadism: Pathogenesis, Diagnosis and Treatment. Nat. Rev. Urol. 2011, 8, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Snyder, P.J.; Bhasin, S.; Cunningham, G.R.; Matsumoto, A.M.; Stephens-Shields, A.J.; Cauley, J.A.; Ellenberg, S.S. Effects of Testosterone Treatment in Older Men. N. Engl. J. Med. 2016, 374, 611–624. [Google Scholar] [CrossRef]
- Wu, F.C.; Tajar, A.; Beynon, J.M.; Pye, S.R.; Silman, A.J.; Finn, J.D.; O’Neill, T.W.; Bartfai, G.; Casanueva, F.F.; Forti, G.; et al. Identification of Late-Onset Hypogonadism in Middle-Aged and Elderly Men. N. Engl. J. Med. 2010, 363, 123–135. [Google Scholar] [CrossRef]
- Basaria, S. Male Hypogonadism. Lancet 2014, 383, 1250–1263. [Google Scholar] [CrossRef]
- Bhattacharya, R.K.; Bhattacharya, S.B. Late-Onset Hypogonadism and Testosterone Replacement in Older Men. Clin. Geriatr. Med. 2015, 31, 631–644. [Google Scholar] [CrossRef]
- Bhasin, S.; Brito, J.P.; Cunningham, G.R.; Hayes, F.J.; Hodis, H.N.; Matsumoto, A.M.; Snyder, P.J.; Swerdloff, R.S.; Wu, F.C.; Yialamas, M.A. Testosterone Therapy in Men with Hypogonadism: An Endocrine Society* Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2018, 103, 1715–1744. [Google Scholar] [CrossRef]
- Ramasamy, R.; Wilken, N.; Scovell, J.M.; Lipshultz, L.I. Effect of Testosterone Supplementation on Symptoms in Men with Hypogonadism. Eur. Urol. 2015, 67, 176–177. [Google Scholar] [CrossRef]
- Zhu, Y.; Ge, J.; Huang, C.; Liu, H.; Jiang, H. Application of Mesenchymal Stem Cell Therapy for Aging Frailty: From Mechanisms to Therapeutics. Theranostics 2021, 11, 5675–5685. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Huang, X.; Dou, L.; Yan, M.; Shen, T.; Tang, W.; Li, J. Aging and Aging-Related Diseases: From Molecular Mechanisms to Interventions and Treatments. Signal Transduct. Target Ther. 2022, 7, 391. [Google Scholar] [CrossRef] [PubMed]
- Giovannelli, L.; Bari, E.; Jommi, C.; Tartara, F.; Armocida, D.; Garbossa, D.; Cofano, F.; Torre, M.L.; Segale, L. Mesenchymal Stem Cell Secretome and Extracellular Vesicles for Neurodegenerative Diseases: Risk-Benefit Profile and Next Steps for the Market Access. Bioact. Mater. 2023, 29, 16–35. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-J.; Wei, R.; Li, F.; Liao, S.-Y.; Tse, H.-F. Mesenchymal Stromal Cell-Derived Exosomes in Cardiac Regeneration and Repair. Stem Cell Rep. 2021, 16, 1662–1673. [Google Scholar] [CrossRef] [PubMed]
- Guillamat-Prats, R. The Role of Msc in Wound Healing, Scarring and Regeneration. Cells 2021, 10, 1729. [Google Scholar] [CrossRef]
- Watanabe, Y.; Tsuchiya, A.; Terai, S. The Development of Mesenchymal Stem Cell Therapy in the Present, and the Perspective of Cell-Free Therapy in the Future. Clin. Mol. Hepatol. 2021, 27, 70–80. [Google Scholar] [CrossRef]
- Yao, X.; Wei, W.; Wang, X.; Li, C.; Björklund, M.; Ouyang, H. Stem Cell Derived Exosomes: MicroRNA Therapy for Age-Related Musculoskeletal Disorders. Biomaterials 2019, 224, 119492. [Google Scholar] [CrossRef]
- Phinney, D.G.; Pittenger, M.F. Concise Review: MSC-Derived Exosomes for Cell-Free Therapy. Stem Cells 2017, 35, 851–858. [Google Scholar] [CrossRef]
- Ha, D.H.; Kim, H.-K.; Lee, J.; Kwon, H.H.; Park, G.-H.; Yang, S.H.; Jung, J.Y.; Choi, H.; Lee, J.H.; Sung, S.; et al. Mesenchymal Stem/Stromal Cell-Derived Exosomes for Immunomodulatory Therapeutics and Skin Regeneration. Cells 2020, 9, 1157. [Google Scholar] [CrossRef]
- Staff, N.P.; Jones, D.T.; Singer, W. Mesenchymal Stromal Cell Therapies for Neurodegenerative Diseases. Mayo Clin. Proc. 2019, 94, 892–905. [Google Scholar] [CrossRef]
- Malekpour, K.; Hazrati, A.; Zahar, M.; Markov, A.; Zekiy, A.O.; Navashenaq, J.G.; Roshangar, L.; Ahmadi, M. The Potential Use of Mesenchymal Stem Cells and Their Derived Exosomes for Orthopedic Diseases Treatment. Stem Cell Rev. Rep. 2022, 18, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Xiao, L.; Peng, J.; Qian, Y.-Q.; Huang, C.-C. Exosomes Derived from Bone Marrow Mesenchymal Stem Cells Improve Osteoporosis through Promoting Osteoblast Proliferation via MAPK Pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3962–3970. [Google Scholar] [PubMed]
- Chen, H.X.; Liang, F.C.; Gu, P.; Xu, B.L.; Xu, H.J.; Wang, W.T.; Hou, J.Y.; Xie, D.X.; Chai, X.Q.; An, S.J. Exosomes Derived from Mesenchymal Stem Cells Repair a Parkinson’s Disease Model by Inducing Autophagy. Cell Death Dis. 2020, 11, 288. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Yin, Z.; Chen, F.; Lei, P. Mesenchymal Stem Cell-Derived Exosome: A Promising Alternative in the Therapy of Alzheimer’s Disease. Alzheimer’s Res. Ther. 2020, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Du, X.; Wang, C.; Zhang, J.; Liu, C.; Li, Y.; Jiang, H. Therapeutic Effects of Human Umbilical Cord Mesenchymal Stem Cell-Derived Microvesicles on Premature Ovarian Insufficiency in Mice. Stem Cell Res. Ther. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Ros, J.; Romero-Garcia, N.; Mas-Bargues, C.; Monleon, D.; Gordevicius, J.; Brooke, R.T.; Dromant, M.; Diaz, A.; Derevyanko, A.; Guio-Carrion, A.; et al. Small Extracellular Vesicles from Young Adipose-Derived Stem Cells Prevent Frailty, Improve Health Span, and Decrease Epigenetic Age in Old Mice. Sci. Adv. 2022, 8, eabq2226. [Google Scholar] [CrossRef] [PubMed]
- Krampera, M.; Le Blanc, K. Mesenchymal Stromal Cells: Putative Microenvironmental Modulators Become Cell Therapy. Cell Stem Cell 2021, 28, 1708–1725. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Luo, M.; Wei, X. Mesenchymal Stem/Stromal Cells in Cancer Therapy. J. Hematol. Oncol. 2021, 14, 195. [Google Scholar] [CrossRef]
- Fernández-Santos, M.E.; Garcia-Arranz, M.; Andreu, E.J.; García-Hernández, A.M.; López-Parra, M.; Villarón, E.; Sepúlveda, P.; Fernández-Avilés, F.; García-Olmo, D.; Prosper, F.; et al. Optimization of Mesenchymal Stromal Cell (Msc) Manufacturing Processes for a Better Therapeutic Outcome. Front. Immunol. 2022, 13, 918565. [Google Scholar] [CrossRef]
- Wragg, N.M.; Tampakis, D.; Stolzing, A. Cryopreservation of Mesenchymal Stem Cells Using Medical Grade Ice Nucleation Inducer. Int. J. Mol. Sci. 2020, 21, 8579. [Google Scholar] [CrossRef]
- Yaghoubi, Y.; Movassaghpour, A.; Zamani, M.; Talebi, M.; Mehdizadeh, A.; Yousefi, M. Human Umbilical Cord Mesenchymal Stem Cells Derived-Exosomes in Diseases Treatment. Life Sci. 2019, 233, 116733. [Google Scholar] [CrossRef] [PubMed]
- Harrell, C.R.; Jovicic, N.; Djonov, V.; Volarevic, V. Therapeutic Use of Mesenchymal Stem Cell-Derived Exosomes: From Basic Science to Clinics. Pharmaceutics 2020, 12, 474. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Li, G.; Li, D.; Huang, W.; Zhang, R.; Zhang, H.; Duan, Y.; Wang, B. HucMSC Derived Exosomes Promote Functional Recovery in Spinal Cord Injury Mice via Attenuating Inflammation. Mater. Sci. Eng. C 2018, 89, 194–204. [Google Scholar] [CrossRef]
- Han, Q.; Wang, S.; Chen, D.; Gan, D.; Wang, T. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Reduce Tendon Injuries via the Mir-27b-3p/Arhgap5/Rhoa Signaling Pathway. Acta Biochim. Biophys. Sin. 2022, 54, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhang, J.; Xu, B.; He, Y.; Liu, W.; Li, J.; Zhang, S.; Lin, X.; Su, D.; Wu, T.; et al. Hucmsc-Derived Exosomes Mitigate the Age-Related Retardation of Fertility in Female Mice. Mol. Ther. 2020, 28, 1200–1213. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Wang, F.; Lai, X.; Dong, L.; Luo, P.; Zhang, S.; Yang, C.; Chen, H.; Ma, Y.; Huang, W.; et al. AAV-Mediated Gene Therapy Produces Fertile Offspring in the Lhcgr-Deficient Mouse Model of Leydig Cell Failure. Cell Rep. Med. 2022, 3, 100792. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Zhu, L.; Shen, H.; Lu, J.; Zou, Q.; Huang, C.; Li, H.; Huang, B. Exosomal Mirna-17-5p Derived from Human Umbilical Cord Mesenchymal Stem Cells Improves Ovarian Function in Premature Ovarian Insufficiency by Regulating Sirt7. Stem Cells 2020, 38, 1137–1148. [Google Scholar] [CrossRef]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, C.; Su, Z.; Chang, Q.; Qiu, Y.; Bei, J.; Gaitas, A.; Xiao, J.; Drelich, A.; Khanipov, K.; et al. Endothelial Exosome Plays a Functional Role during Rickettsial Infection. mBio 2021, 12, 10–1128. [Google Scholar] [CrossRef]
- Street, J.M.; Koritzinsky, E.H.; Glispie, D.M.; Yuen, P.S.T. Urine Exosome Isolation and Characterization. Methods Mol. Biol. 2017, 1641, 413–423. [Google Scholar]
- Faruqu, F.N.; Xu, L.; Al-Jamal, K.T. Preparation of Exosomes for Sirna Delivery to Cancer Cells. J. Vis. Exp. 2018, 142, e58814. [Google Scholar]
- Sabzian-Melei, R.; Zare-Shahneh, A.; Zhandi, M.; Yousefi, A.R.; Rafieian-Naeini, H.R. Effects of Dietary Supplementation of Different Sources and Levels of Selenium on the Semen Quality and Reproductive Performance in Aged Broiler Breeder Roosters. Poult. Sci. 2022, 101, 101908. [Google Scholar] [CrossRef] [PubMed]
- Misiakiewicz, K.; Kolasa, A.; Kondarewicz, A.; Marchlewicz, M.; Wiszniewska, B. Expression of the c-Kit Receptor in Germ Cells of the Seminiferous Epithelium in Rats with Hormonal Imbalance. Reprod. Biol. 2013, 13, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Shan, G.; Jahangiri, S.; Chen, W.; Gholamhoseini, B.; Zhang, Z.; Moskovtsev, S.; Librach, C.; Jarvi, K.; Sun, Y. Staining-free, Automated Sperm Analysis for In Vitro Fertilization Lab Use. J. Urol. 2022, 208, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Ma, Y.; Feng, X.; Deng, R.; Ke, Q.; Xiang, A.P.; Deng, C. Endosialin Defines Human Stem Leydig Cells with Regenerative Potential. Hum. Reprod. 2020, 35, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Nakata, H.; Kadomoto, S.; Iwamoto, H.; Yaegashi, H.; Iijima, M.; Kawaguchi, S.; Nohara, T.; Shigehara, K.; Izumi, K.; et al. Three-Dimensional Morphological Analysis of Spermatogenesis in Aged Mouse Testes. Sci. Rep. 2021, 11, 23007. [Google Scholar] [CrossRef]
- Huang, Y.; Li, X.; Sun, X.; Yao, J.; Gao, F.; Wang, Z.; Hu, J.; Wang, Z.; Ouyang, B.; Tu, X.; et al. Anatomical Transcriptome Atlas of the Male Mouse Reproductive System during Aging. Front. Cell Dev. Biol. 2022, 9, 782824. [Google Scholar] [CrossRef]
- Arabpour, M.; Saghazadeh, A.; Rezaei, N. Anti-Inflammatory and M2 Macrophage Polarization-Promoting Effect of Mesenchymal Stem Cell-Derived Exosomes. Int. Immunopharmacol. 2021, 97, 107823. [Google Scholar] [CrossRef]
- Harrell, C.R.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Mesenchymal Stem Cell-Derived Exosomes and Other Extracellular Vesicles as New Remedies in the Therapy of Inflammatory Diseases. Cells 2019, 8, 1605. [Google Scholar] [CrossRef]
- Santiago, J.; Silva, J.V.; Alves, M.G.; Oliveira, P.F.; Fardilha, M. Testicular Aging: An Overview of Ultrastructural, Cellular, and Molecular Alterations. J. Gerontol. A Biol. Sci. Med. Sci. 2019, 74, 860–871. [Google Scholar] [CrossRef]
- Dong, S.; Chen, C.; Zhang, J.; Gao, Y.; Zeng, X.; Zhang, X. Testicular Aging, Male Fertility and Beyond. Front. Endocrinol. 2022, 13, 1012119. [Google Scholar] [CrossRef] [PubMed]
- Basisty, N.; Kale, A.; Jeon, O.H.; Kuehnemann, C.; Payne, T.; Rao, C.; Holtz, A.; Shah, S.; Sharma, V.; Ferrucci, L.; et al. A Proteomic Atlas of Senescence-Associated Secretomes for Aging Biomarker Development. PLoS Biol. 2020, 18, e3000599. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; di Fagagna, F.D. Cellular Senescence in Ageing: From Mechanisms to Therapeutic Opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Matias, I.; Diniz, L.P.; Damico, I.V.; Araujo, A.P.B.; Neves, L.D.S.; Vargas, G.; Leite, R.E.P.; Suemoto, C.K.; Nitrini, R.; Jacob-Filho, W.; et al. Loss of Lamin-B1 and Defective Nuclear Morphology Are Hallmarks of Astrocyte Senescence in Vitro and in the Aging Human Hippocampus. Aging Cell 2022, 21, e13521. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; An, Y.; He, J.; Huang, B.; Zhu, J.; Gao, M.; Zhang, S.; Wang, X.; Yang, B.; Xie, Z. Berberine Ameliorates Cellular Senescence and Extends the Lifespan of Mice via Regulating p16 and Cyclin Protein Expression. Aging Cell 2020, 19, e13060. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Magana, A.; Blanco, F.J. Human PCNA Structure, Function and Interactions. Biomolecules 2020, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Aihara, H. DNA Ligase and PCNA: Double-Ring Down to Seal a Break in DNA. Structure 2022, 30, 324–326. [Google Scholar] [CrossRef]
- Xie, T.; Li, L. Stem Cells and Their Niche: An Inseparable Relationship. Development 2007, 134, 2001–2006. [Google Scholar] [CrossRef]
- Kim, J.Y.; Jung, H.J.; Yoon, M.J. Vasa (Ddx4) Is a Putative Marker for Spermatogonia, Spermatocytes and Round Spermatids in Stallions. Reprod. Domest. Anim. 2015, 50, 1032–1038. [Google Scholar] [CrossRef]
- Majhi, R.K.; Kumar, A.; Yadav, M.; Kumar, P.; Maity, A.; Giri, S.C.; Goswami, C. Light and Electron Microscopic Study of Mature Spermatozoa from White Pekin Duck (Anas platyrhynchos): An Ultrastructural and Molecular Analysis. Andrology 2016, 4, 232–244. [Google Scholar] [CrossRef]
- Winnall, W.R.; Hedger, M.P. Phenotypic and Functional Heterogeneity of the Testicular Macrophage Population: A New Regulatory Model. J. Reprod. Immunol. 2013, 97, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, S.; Meinhardt, A. The Macrophages in Testis Function. J. Reprod. Immunol. 2017, 119, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, S.; Jiang, S.; Huang, Z.; Chen, X.; Guo, H.; Li, M.; Zheng, S. Evaluation of Immune Status in Testis and Macrophage Polarization Associated with Testicular Damage in Patients with Nonobstructive Azoospermia. Am. J. Reprod. Immunol. 2021, 86, e13481. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Huang, W.; Liu, S.; Cai, S.; Hong, L.; Xiao, W.; Thiele, K.; Zeng, Y.; Song, M.; Diao, L. Impacts of Immunometabolism on Male Reproduction. Front. Immunol. 2021, 12, 658432. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; Pang, M.-G. Mitochondrial Functionality in Male Fertility: From Spermatogenesis to Fertilization. Antioxidants 2021, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xiao, H.; Zhang, X.; Qiukai, E.; Gong, X.; Li, T.; Zhang, X. Decreased Microrna-125b-5p Disrupts Follicle Steroidogenesis through Targeting Pak3/Erk1/2 Signalling in Mouse Preantral Follicles. Metab. Clin. Exp. 2020, 107, 154241. [Google Scholar] [CrossRef] [PubMed]
- Presslauer, C.; Bizuayehu, T.T.; Kopp, M.; Fernandes, J.M.O.; Babiak, I. Dynamics of miRNA Transcriptome during Gonadal Development of Zebrafish. Sci. Rep. 2017, 7, srep43850. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, H.; Mei, J.; Cao, Z.; Tang, Y.; Huang, R.; Huang, Y. Sertoli Cell-Derived Exosome-Mediated Transfer of Mir-145-5p Inhibits Leydig Cell Steroidogenesis by Targeting Steroidogenic Factor 1. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2021, 35, e21660. [Google Scholar] [CrossRef]
- Gao, G.-Z.; Zhao, Y.; Li, H.-X.; Li, W. Bisphenol A-Elicited miR-146a-5p Impairs Murine Testicular Steroidogenesis through Negative Regulation of Mta3 Signaling. Biochem. Biophys. Res. Commun. 2018, 501, 478–485. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, L.; Li, H.; Zhao, X.; Ding, Y.; Yao, Y.; Wang, L. Association between Yili Goose Sperm Motility and Expression Profiles of Mrna and Mirna in Testis. BMC Genom. 2023, 24, 640. [Google Scholar] [CrossRef]
- Larriba, S.; Sánchez-Herrero, J.F.; Pluvinet, R.; López-Rodrigo, O.; Bassas, L.; Sumoy, L. Seminal Extracellular Vesicle Sncrna Sequencing Reveals Altered Mirna/Isomir Profiles as Sperm Retrieval Biomarkers for Azoospermia. Andrology 2023, 12, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, L.; Zhou, Y.; Tian, J.; Li, Y.; Xie, Z.; Pei, C.; Yan, B.; Ma, L. The Role of miR-199a-3p in Inhibiting the Proliferation of Spermatogonial Stem Cells under Heat Stress. Theriogenology 2023, 211, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Maresch, C.C.; Petry, S.F.; Paradowska-Dogan, A.; Bhushan, S.; Chang, Y.; Wrenzycki, C.; Schuppe, H.-C.; Houska, P.; Hartmann, M.F.; et al. Elevated CCL2 Causes Leydig Cell Malfunction in Metabolic Syndrome. J. Clin. Investig. 2020, 5, e134882. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xie, Y.; Chen, H.; Lv, L.; Yao, J.; Zhang, M.; Xia, K.; Feng, X.; Li, Y.; Liang, X.; et al. FOXO4-DRI Alleviates Age-Related Testosterone Secretion Insufficiency by Targeting Senescent Leydig Cells in Aged Mice. Aging 2020, 12, 1272–1284. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, F.; Ye, L.; Zirkin, B.; Chen, H. Steroidogenesis in Leydig Cells: Effects of Aging and Environmental Factors. Reproduction 2017, 154, R111–R122. [Google Scholar] [CrossRef]
- Gao, F.; Li, G.; Liu, C.; Gao, H.; Wang, H.; Liu, W.; Chen, M.; Shang, Y.; Wang, L.; Shi, J.; et al. Autophagy Regulates Testosterone Synthesis by Facilitating Cholesterol Uptake in Leydig Cells. J. Cell Biol. 2018, 217, 2103–2119. [Google Scholar] [CrossRef]
- Ding, H.; Zhao, J.; Liu, H.; Wang, J.; Lu, W. BMAL1 Knockdown Promoted Apoptosis and Reduced Testosterone Secretion in TM3 Leydig Cell Line. Gene 2020, 747, 144672. [Google Scholar] [CrossRef]
- Lai, R.C.; Yeo, R.W.Y.; Tan, K.H.; Lim, S.K. Mesenchymal Stem Cell Exosome Ameliorates Reperfusion Injury Through Proteomic Complementation. Regen. Med. 2013, 8, 197–209. [Google Scholar] [CrossRef]
- Panfoli, I.; Ravera, S.; Podestà, M.; Cossu, C.; Santucci, L.; Bartolucci, M.; Bruschi, M.; Calzia, D.; Sabatini, F.; Bruschettini, M.; et al. Exosomes from Human Mesenchymal Stem Cells Conduct Aerobic Metabolism in Term and Preterm Newborn Infants. FASEB J. 2016, 30, 1416–1424. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, P.; Chen, X.; Gao, F.; Xiang, A.P.; Deng, C.; Xia, K.; Gao, Y. Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosomes Rescue Testicular Aging. Biomedicines 2024, 12, 98. https://doi.org/10.3390/biomedicines12010098
Luo P, Chen X, Gao F, Xiang AP, Deng C, Xia K, Gao Y. Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosomes Rescue Testicular Aging. Biomedicines. 2024; 12(1):98. https://doi.org/10.3390/biomedicines12010098
Chicago/Turabian StyleLuo, Peng, Xuren Chen, Feng Gao, Andy Peng Xiang, Chunhua Deng, Kai Xia, and Yong Gao. 2024. "Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosomes Rescue Testicular Aging" Biomedicines 12, no. 1: 98. https://doi.org/10.3390/biomedicines12010098