1. Introduction
Glycososaminoglycans (GAGs) are a class of linear anionic periodic polysaccharides that are made up of repetitive disaccharide building blocks containing a uronic acid (glucuronic, GlcA or iduronic, IdoA) and a hexosamine (N-Acetylglycosamide, GlcNAc or N-Acetylgalactososamide GalNAc) [
1]. GAGs are located in the extracellular matrix of the cell, where they participate in many key biochemical processes such as angiogenesis, anticoagulation, cellular communication and adhesion [
2,
3]. Their involvement in these processes is mediated through direct interactions with diverse protein targets such as collagens [
4], chemokines [
5] and growth factors [
6,
7]. All this makes GAGs to be essential players in a number of diseases and disorders including cancer [
8], Alzheimer’s [
9] and Parkinson’s disease [
10], autoimmune diseases [
11] and arthritis [
12]. Therefore, GAGs are very promising potential molecular targets for novel regenerative medicine strategies [
13,
14,
15]. Chemically, GAGs are immensely heterogeneous. Depending on their disaccharide unit composition, glycosidic linkage and sulfation pattern, they are classified into several groups: hyaluronic acid (HA), chondroitin sulfate/dermatan sulfate (CS/DS), heparin/heparan sulfate (HP/HS) and keratan sulfate (KS). Altogether, 202 different GAG disaccharide variants in mammals are known [
3]. Such heterogeneity, which could be also present within the same GAG chain, as well as GAG’s high flexibility and periodicity, renders these molecules profoundly challenging to analyse using the experimental techniques only [
16]. Therefore, computational approaches could be particularly efficient in the GAG research [
17,
18]. Recently, many interdisciplinary studies proved that a combination of the experimental and theoretical approaches could be especially promising in studying biomolecular systems containing GAGs [
19,
20]. At the same time, for computational researchers, there are still many challenges related to the physico-chemical properties of GAGs to overcome. They include their anionic nature, which makes it essential to use appropriate treatment of the electrostatics, ions and solvent, which is much more abundant in protein-GAG interfaces than in the complexes of proteins with other classes of biomolecules [
21]. GAGs periodicity can result in multipose binding, in which several protein-GAG complex configurations can have similar free binding energies and, therefore, co-exist [
22]. Finally, one of the key challenges in understanding GAG molecular interactions relates to deciphering the “sulfation code” [
23], which should assist in the explanation and prediction of GAG specificity [
24,
25]. A recent work of Holmes et al. can be considered as a breakthrough in terms of understanding the “sulfation code” [
26]. In this work, a number of molecular descriptors of several HS variants were analysed with the molecular dynamics (MD) approach, and it was shown that a combination of these partially interdependent MD-derived parameters determine the conformational behaviour and binding propensities of the GAGs, while the data about the GAG sequence alone are not sufficient to improve the knowledge on these complex molecules in terms of the “sulfation code”.
In the present study, we aim to contribute to the comprehension of the “sulfation code” by applying MD-based analysis, which allows for essential advances in understanding GAG properties [
27], to CS and DS molecules. CS and DS are very similar GAG molecules made up of GalNAc(
)GlcA(
) and GalNAc(
)IdoA(
) disaccharide units, respectively [
28]. CS and DS chains are usually composed of 40 to 100 disaccharide units [
29]. CS can be found in the cartilage and is involved in the bone resorption process [
30], while DS can be found both in the skin, blood vessels and lungs and has antithrombotic activity [
31]. When sulfated in the 4th of the 6th position of GalNAc, CS has a charge of −2 per disaccharide unit (CS4 and CS6, respectively). DS has predominantly sulfation in the 4th position and its disaccharide unit has also a net charge of −2 [
32]. Sometimes, GlcA and IdoA can undergo epimerisation, and then a GAG chain has both CS and DS parts. Despite their similarities, CS and DS have very distinct protein binding properties: in particular, there is experimental evidence so far suggesting that if there are complexes with DS, the same proteins also bind other GAGs, while there are many indications of the CS binding specificity, meaning that some protein can bind only CS but not DS [
3]. It was also shown, both with NMR and molecular dynamics (MD)-base techniques that CS4 and CS6 can bind with significantly different affinities to the same proteins [
33] suggesting that it is not exclusively a net charge that drives these interactions. MD simulations have also proved to be successful in reproducing experimental data for unbound CS oligosaccharides. Already in 1999, Kaufmann et al. performed 4 ns MD simulations of the CS4 tetrasaccharide to characterise its glycosidic linkage conformational space, hydration properties and H-bonding [
34]. It was shown that in comparison to non-sulfated oligosaccharides, CS is highly hydrated thanks to its negative charge. Interestingly, this hydration partially remains high also when they bind to proteins [
21]. Samantray et al. characterised the dynamics of several GAG disaccharides including CS4 and CS6 and established the dependence of their behaviour on the salt presence and type [
35]. Guvench’s group constructed atomic models of CS/DS oligosaccharides, rigorously analysed their conformational space and established the dependence of their conformational ensembles on the interactions with Ca
and their sulfation pattern [
36,
37,
38,
39,
40]. Recently, MD simulations with CHARMM and GLYCAM06 force fields were used to characterise all possible disaccharide variants of CS in terms of their free energy landscape [
41,
42]. In the latter publication, the authors performed a rigorous analysis of the disaccharide torsional space, intra- and intermolecular H-bonds, bridging water molecules and principal components of movements. They conclude that the observed distinct conformational dynamism in different CS disaccharides is the reason that unique electrostatic surfaces exist that could be a key for protein recognition.
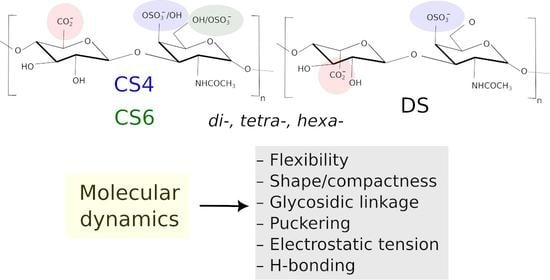
In this work, we compare the properties of CS4, CS6 and DS of different lengths (di-, tetra- and hexasaccharides—dp2, dp4 and dp6, where dp stays for the degree of polymerisation) in two chain variants (Set 1 and Set 2) differing from each other by the type of the residue at the reducing and non-reducing ends of the oligosaccharides (
Figure 1).
In total, 18 oligosaccharides are analysed and compared to dissect the effect of the sulfation and the type of uronic acid on the dynamic and conformational properties of these GAGs, which should, in turn, determine the differences in their binding specificity. In particular, flexibility defined in terms of the atomic fluctuations, the radius of gyration, end-to-end distance, molecular volume, interactions with the ions and solvent, internal hydrogen bonds, glycosidic linkage and ring puckering conformations have been used as molecular descriptors similarly to other GAG conformational studies [
26]. We find that the differences between CS4, CS6 and DS can be established by considering a combination of the analysed dynamic parameters. Our data are in line with the results obtained for 3-O-sulfated HS variants by Holmes et al. suggesting that the GAG “sulfation code” is a concept which understanding should be established based on the data obtained from the dynamics of these systems [
26]. In these terms, our work adds to the general knowledge about the “sulfation code”, which could be of high relevance for the novel approaches in the GAG-related drug development for a number of diseases where these molecules are mediators of the underlying molecular processes.
4. Conclusions
We applied MD-based analysis with the aim to understand the differences between CS4, CS6 and DS oligosaccharides in terms of several structural/dynamical descriptors (RMSD, R, EED, molecular volume, glycosidic linkages and puckering conformational space, internal electrostatic tension, interactions with counterions and H-bonding propensities). The descriptors are partially interdependent and could, therefore, indicate similar trends. At the same time, some of them are able to reveal significant differences between the analysed molecules, which are indistinguishable from other descriptors. Such significant differences have been observed for the flexibility of the studied GAG types, suggesting DS to be more rigid than both CSs, while, at the same time, the puckering space of IdoA, which is one of the residues within the disaccharide repetitive unit of DS, is essentially broader than the one of GlcA, a residue in both CSs instead. The conformational ensemble of DS could be divided into two major groups of conformations, for which the exchange is relatively slow, while CSs conformations represent a single major population exchanging rapidly with several minor conformational populations. In terms of shape, DS is the most compact, while CS6 is the most elongated molecule. Electrostatic properties and H-bonding propensities for all three GAGs are essentially different: CS6 has the highest internal electrostatic tension, while DS has the least intramolecular H-bond acceptors that are available for establishing an H-bond upon binding to a GAG binding partner. These results suggest principal differences in the kinetic and thermodynamic patterns of the intermolecular interactions that can be established by the three GAG types with proteins and other biological binding partners. In particular, the in silico obtained data for these GAGs in terms of their potential protein binding specificity can be interpreted as follows. 1. The sulfation in position 6 contributes to the higher flexibility of the GAG in comparison to the sulfation in position 4 and, so could favouritise a higher diversity of its possible conformations upon binding, leading to the lower entropic loss upon the formation of a protein-GAG complex and, therefore, higher affinity. At the same time, this higher flexibility of the CS6 could also result in the binding poses being more specific than in the case of CS4 and DS. 2. There are fewer available H-bond acceptors in the unbound CS than in the DS meaning that there are more putatively specific patterns of the H-bonding for CS to be expected when bound to the protein. 3. Finally, a more flexible IdoA ring puckering conformational ensemble in DS in comparison to the GlcA one in CS could be the reason for the different entropic patterns of these GAGs in the binding. Systematic and rigorous experiments should be performed to find out which thermodynamic trends qualitatively suggested by our data, are dominant in the protein-GAG binding. Isothermal titration calorimetry could potentially be able to answer this question and verify our findings. To summarise, this study represents a systematic step towards understanding the molecular basis of the GAG “sulfation code” and its dynamic nature, which could be of high potential interest for the GAG-based drug design in the field of regenerative medicine.